| Full name: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | Alias Symbol: PI3K | ||
| Type: protein-coding gene | Cytoband: 3q26.32 | ||
| Entrez ID: 5290 | HGNC ID: HGNC:8975 | Ensembl Gene: ENSG00000121879 | OMIM ID: 171834 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PIK3CA involved pathways:
Expression of PIK3CA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIK3CA | 5290 | 204369_at | 0.1489 | 0.7283 | |
| GSE20347 | PIK3CA | 5290 | 204369_at | 0.5354 | 0.0051 | |
| GSE23400 | PIK3CA | 5290 | 204369_at | 0.6435 | 0.0000 | |
| GSE26886 | PIK3CA | 5290 | 204369_at | 0.4954 | 0.2207 | |
| GSE29001 | PIK3CA | 5290 | 204369_at | 0.7311 | 0.1538 | |
| GSE38129 | PIK3CA | 5290 | 204369_at | 0.6421 | 0.0000 | |
| GSE45670 | PIK3CA | 5290 | 204369_at | 0.1383 | 0.5461 | |
| GSE53622 | PIK3CA | 5290 | 61893 | 0.5868 | 0.0000 | |
| GSE53624 | PIK3CA | 5290 | 61893 | 0.5532 | 0.0000 | |
| GSE63941 | PIK3CA | 5290 | 204369_at | 0.3733 | 0.5925 | |
| GSE77861 | PIK3CA | 5290 | 204369_at | 0.8179 | 0.0189 | |
| GSE97050 | PIK3CA | 5290 | A_23_P92057 | 0.1813 | 0.3871 | |
| SRP007169 | PIK3CA | 5290 | RNAseq | 1.9627 | 0.0026 | |
| SRP008496 | PIK3CA | 5290 | RNAseq | 1.6044 | 0.0001 | |
| SRP064894 | PIK3CA | 5290 | RNAseq | 0.4752 | 0.0130 | |
| SRP133303 | PIK3CA | 5290 | RNAseq | 0.8617 | 0.0000 | |
| SRP159526 | PIK3CA | 5290 | RNAseq | 0.9277 | 0.0080 | |
| SRP193095 | PIK3CA | 5290 | RNAseq | 0.6278 | 0.0000 | |
| SRP219564 | PIK3CA | 5290 | RNAseq | 0.0193 | 0.9529 | |
| TCGA | PIK3CA | 5290 | RNAseq | 0.1442 | 0.0158 |
Upregulated datasets: 2; Downregulated datasets: 0.
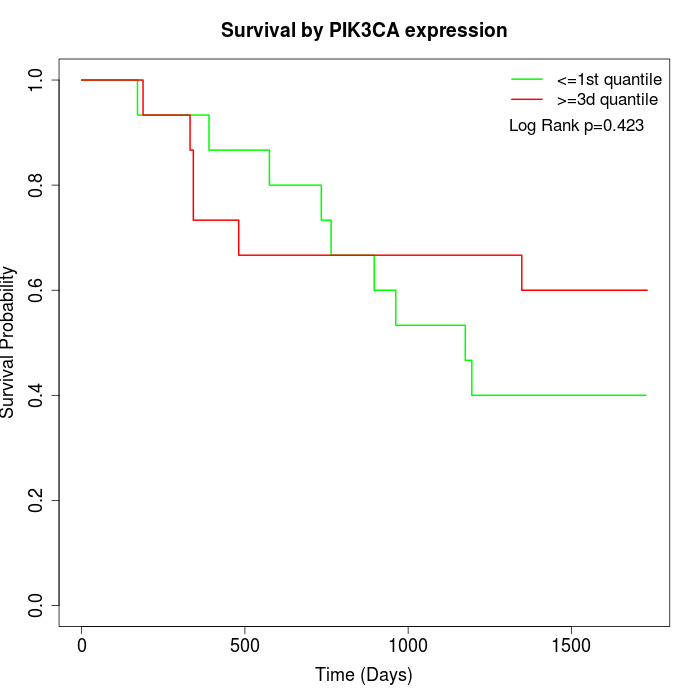
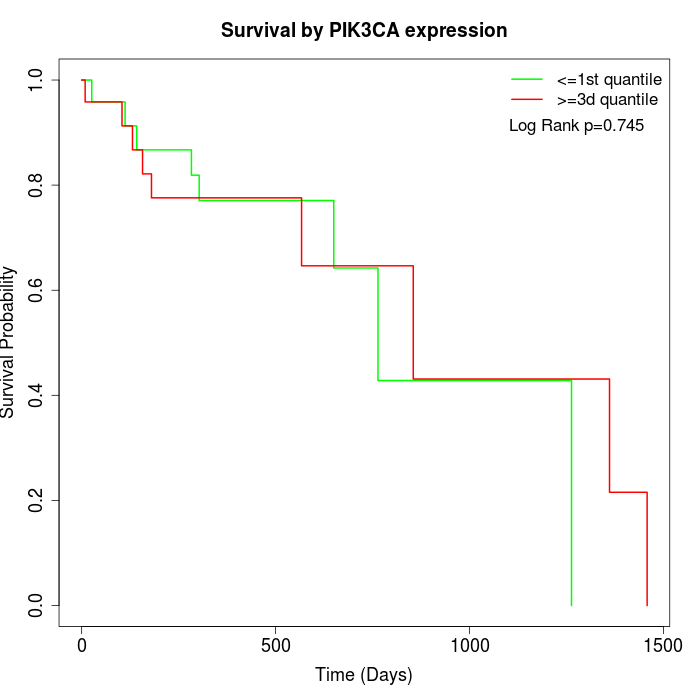
Survival by PIK3CA expression:
Note: Click image to view full size file.
Copy number change of PIK3CA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIK3CA | 5290 | 25 | 0 | 5 | |
| GSE20123 | PIK3CA | 5290 | 25 | 0 | 5 | |
| GSE43470 | PIK3CA | 5290 | 24 | 0 | 19 | |
| GSE46452 | PIK3CA | 5290 | 19 | 1 | 39 | |
| GSE47630 | PIK3CA | 5290 | 27 | 2 | 11 | |
| GSE54993 | PIK3CA | 5290 | 1 | 18 | 51 | |
| GSE54994 | PIK3CA | 5290 | 41 | 0 | 12 | |
| GSE60625 | PIK3CA | 5290 | 0 | 6 | 5 | |
| GSE74703 | PIK3CA | 5290 | 20 | 0 | 16 | |
| GSE74704 | PIK3CA | 5290 | 16 | 0 | 4 | |
| TCGA | PIK3CA | 5290 | 78 | 0 | 18 |
Total number of gains: 276; Total number of losses: 27; Total Number of normals: 185.
Somatic mutations of PIK3CA:
Generating mutation plots.
Highly correlated genes for PIK3CA:
Showing top 20/1253 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIK3CA | CCNT1 | 0.800525 | 3 | 0 | 3 |
| PIK3CA | SMCR8 | 0.74885 | 3 | 0 | 3 |
| PIK3CA | UHMK1 | 0.737559 | 4 | 0 | 4 |
| PIK3CA | FXR1 | 0.734625 | 10 | 0 | 10 |
| PIK3CA | REV1 | 0.732879 | 3 | 0 | 3 |
| PIK3CA | ZNF32 | 0.730821 | 5 | 0 | 5 |
| PIK3CA | METAP1D | 0.723082 | 4 | 0 | 3 |
| PIK3CA | PTPMT1 | 0.714727 | 3 | 0 | 3 |
| PIK3CA | RGMB | 0.711549 | 3 | 0 | 3 |
| PIK3CA | SNX30 | 0.711135 | 3 | 0 | 3 |
| PIK3CA | CBFA2T2 | 0.710999 | 4 | 0 | 3 |
| PIK3CA | UCN | 0.710268 | 3 | 0 | 3 |
| PIK3CA | PCGF6 | 0.70781 | 5 | 0 | 4 |
| PIK3CA | ALMS1 | 0.700313 | 3 | 0 | 3 |
| PIK3CA | USP20 | 0.699863 | 3 | 0 | 3 |
| PIK3CA | LRRC34 | 0.697827 | 3 | 0 | 3 |
| PIK3CA | CDKL3 | 0.697441 | 3 | 0 | 3 |
| PIK3CA | ZBED1 | 0.695778 | 3 | 0 | 3 |
| PIK3CA | USP3 | 0.695546 | 3 | 0 | 3 |
| PIK3CA | ZNF322 | 0.690679 | 7 | 0 | 7 |
For details and further investigation, click here