| Full name: pitrilysin metallopeptidase 1 | Alias Symbol: MP1|KIAA1104|hMP1|PreP | ||
| Type: protein-coding gene | Cytoband: 10p15.2 | ||
| Entrez ID: 10531 | HGNC ID: HGNC:17663 | Ensembl Gene: ENSG00000107959 | OMIM ID: 618211 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PITRM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PITRM1 | 10531 | 205273_s_at | 0.1839 | 0.6991 | |
| GSE20347 | PITRM1 | 10531 | 205273_s_at | 0.0372 | 0.8454 | |
| GSE23400 | PITRM1 | 10531 | 205273_s_at | 0.1806 | 0.0355 | |
| GSE26886 | PITRM1 | 10531 | 205273_s_at | -0.5434 | 0.0508 | |
| GSE29001 | PITRM1 | 10531 | 205273_s_at | 0.0685 | 0.8555 | |
| GSE38129 | PITRM1 | 10531 | 205273_s_at | 0.0381 | 0.7983 | |
| GSE45670 | PITRM1 | 10531 | 205273_s_at | 0.2298 | 0.1709 | |
| GSE53622 | PITRM1 | 10531 | 26245 | 0.0712 | 0.4084 | |
| GSE53624 | PITRM1 | 10531 | 26245 | 0.1721 | 0.0022 | |
| GSE63941 | PITRM1 | 10531 | 205273_s_at | -0.5282 | 0.5336 | |
| GSE77861 | PITRM1 | 10531 | 205273_s_at | -0.1822 | 0.4578 | |
| GSE97050 | PITRM1 | 10531 | A_23_P52402 | 0.2749 | 0.2878 | |
| SRP007169 | PITRM1 | 10531 | RNAseq | 0.1631 | 0.7429 | |
| SRP008496 | PITRM1 | 10531 | RNAseq | 0.0371 | 0.9081 | |
| SRP064894 | PITRM1 | 10531 | RNAseq | 0.1437 | 0.3752 | |
| SRP133303 | PITRM1 | 10531 | RNAseq | -0.0453 | 0.8188 | |
| SRP159526 | PITRM1 | 10531 | RNAseq | -0.1650 | 0.6404 | |
| SRP193095 | PITRM1 | 10531 | RNAseq | -0.0180 | 0.8690 | |
| SRP219564 | PITRM1 | 10531 | RNAseq | -0.2640 | 0.3800 | |
| TCGA | PITRM1 | 10531 | RNAseq | -0.0300 | 0.5377 |
Upregulated datasets: 0; Downregulated datasets: 0.
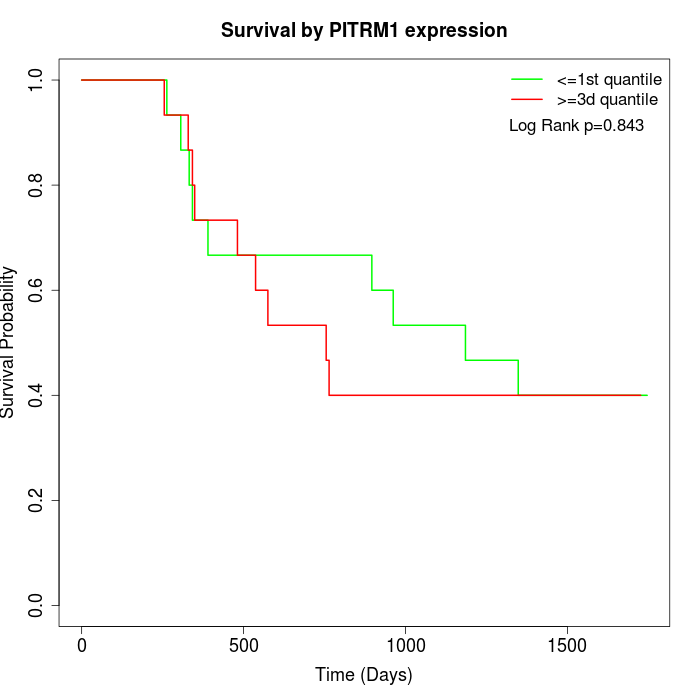
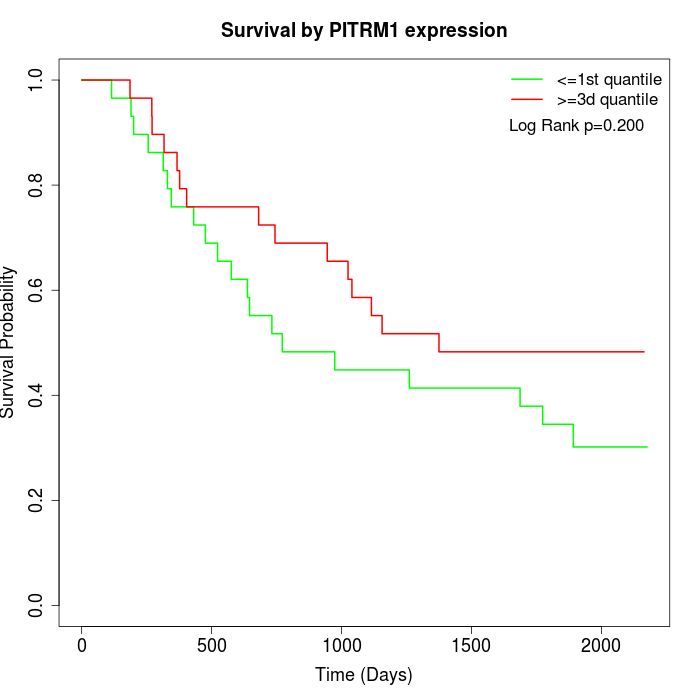
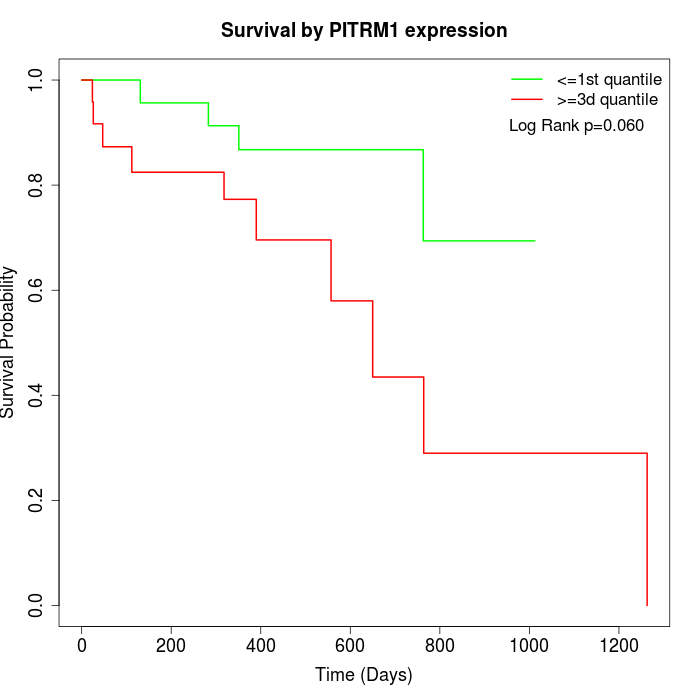
Survival by PITRM1 expression:
Note: Click image to view full size file.
Copy number change of PITRM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PITRM1 | 10531 | 5 | 8 | 17 | |
| GSE20123 | PITRM1 | 10531 | 5 | 7 | 18 | |
| GSE43470 | PITRM1 | 10531 | 3 | 6 | 34 | |
| GSE46452 | PITRM1 | 10531 | 2 | 14 | 43 | |
| GSE47630 | PITRM1 | 10531 | 6 | 14 | 20 | |
| GSE54993 | PITRM1 | 10531 | 11 | 0 | 59 | |
| GSE54994 | PITRM1 | 10531 | 4 | 10 | 39 | |
| GSE60625 | PITRM1 | 10531 | 0 | 0 | 11 | |
| GSE74703 | PITRM1 | 10531 | 2 | 4 | 30 | |
| GSE74704 | PITRM1 | 10531 | 1 | 6 | 13 | |
| TCGA | PITRM1 | 10531 | 18 | 27 | 51 |
Total number of gains: 57; Total number of losses: 96; Total Number of normals: 335.
Somatic mutations of PITRM1:
Generating mutation plots.
Highly correlated genes for PITRM1:
Showing top 20/242 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PITRM1 | ATG3 | 0.776827 | 3 | 0 | 3 |
| PITRM1 | GTF2H5 | 0.756119 | 3 | 0 | 3 |
| PITRM1 | DMTF1 | 0.751074 | 3 | 0 | 3 |
| PITRM1 | RLIM | 0.747458 | 3 | 0 | 3 |
| PITRM1 | GOT1 | 0.746896 | 3 | 0 | 3 |
| PITRM1 | ARFGEF2 | 0.746246 | 3 | 0 | 3 |
| PITRM1 | ZYG11A | 0.731016 | 3 | 0 | 3 |
| PITRM1 | CLK2 | 0.729638 | 3 | 0 | 3 |
| PITRM1 | ZBTB11 | 0.727716 | 3 | 0 | 3 |
| PITRM1 | EIF3A | 0.727518 | 3 | 0 | 3 |
| PITRM1 | LARP1B | 0.725214 | 3 | 0 | 3 |
| PITRM1 | ABCD4 | 0.724073 | 3 | 0 | 3 |
| PITRM1 | ZNF300 | 0.723545 | 3 | 0 | 3 |
| PITRM1 | OMA1 | 0.72354 | 3 | 0 | 3 |
| PITRM1 | TADA1 | 0.722338 | 3 | 0 | 3 |
| PITRM1 | NDUFA10 | 0.72058 | 3 | 0 | 3 |
| PITRM1 | ILF2 | 0.719303 | 3 | 0 | 3 |
| PITRM1 | WWP1 | 0.716386 | 4 | 0 | 3 |
| PITRM1 | TFDP2 | 0.716029 | 3 | 0 | 3 |
| PITRM1 | USP16 | 0.713281 | 3 | 0 | 3 |
For details and further investigation, click here