| Full name: PKHD1 like 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 8q23.1-q23.2 | ||
| Entrez ID: 93035 | HGNC ID: HGNC:20313 | Ensembl Gene: ENSG00000205038 | OMIM ID: 607843 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PKHD1L1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PKHD1L1 | 93035 | 230673_at | -0.7452 | 0.1218 | |
| GSE26886 | PKHD1L1 | 93035 | 230673_at | 0.3104 | 0.0738 | |
| GSE45670 | PKHD1L1 | 93035 | 230673_at | -0.7668 | 0.0000 | |
| GSE63941 | PKHD1L1 | 93035 | 230673_at | 0.1520 | 0.3327 | |
| GSE77861 | PKHD1L1 | 93035 | 230673_at | 0.0812 | 0.2455 | |
| SRP064894 | PKHD1L1 | 93035 | RNAseq | -1.6111 | 0.0024 | |
| SRP133303 | PKHD1L1 | 93035 | RNAseq | -2.5756 | 0.0000 | |
| SRP159526 | PKHD1L1 | 93035 | RNAseq | -1.1256 | 0.2228 | |
| SRP219564 | PKHD1L1 | 93035 | RNAseq | -2.4282 | 0.0005 | |
| TCGA | PKHD1L1 | 93035 | RNAseq | -2.9969 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
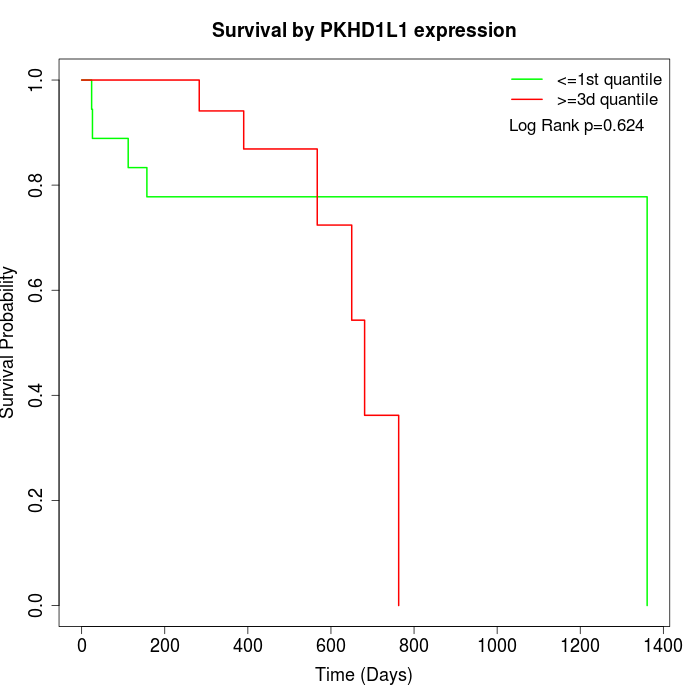
Survival by PKHD1L1 expression:
Note: Click image to view full size file.
Copy number change of PKHD1L1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PKHD1L1 | 93035 | 16 | 0 | 14 | |
| GSE20123 | PKHD1L1 | 93035 | 17 | 0 | 13 | |
| GSE43470 | PKHD1L1 | 93035 | 23 | 0 | 20 | |
| GSE46452 | PKHD1L1 | 93035 | 25 | 0 | 34 | |
| GSE47630 | PKHD1L1 | 93035 | 24 | 0 | 16 | |
| GSE54993 | PKHD1L1 | 93035 | 0 | 20 | 50 | |
| GSE54994 | PKHD1L1 | 93035 | 36 | 1 | 16 | |
| GSE60625 | PKHD1L1 | 93035 | 0 | 4 | 7 | |
| GSE74703 | PKHD1L1 | 93035 | 20 | 0 | 16 | |
| GSE74704 | PKHD1L1 | 93035 | 11 | 0 | 9 | |
| TCGA | PKHD1L1 | 93035 | 60 | 1 | 35 |
Total number of gains: 232; Total number of losses: 26; Total Number of normals: 230.
Somatic mutations of PKHD1L1:
Generating mutation plots.
Highly correlated genes for PKHD1L1:
Showing top 20/49 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PKHD1L1 | IP6K3 | 0.798455 | 3 | 0 | 3 |
| PKHD1L1 | TESC | 0.772867 | 3 | 0 | 3 |
| PKHD1L1 | IGSF10 | 0.741212 | 3 | 0 | 3 |
| PKHD1L1 | SNED1 | 0.73887 | 3 | 0 | 3 |
| PKHD1L1 | KCTD12 | 0.693535 | 3 | 0 | 3 |
| PKHD1L1 | ITIH5 | 0.689303 | 3 | 0 | 3 |
| PKHD1L1 | CLDN5 | 0.681982 | 3 | 0 | 3 |
| PKHD1L1 | S1PR1 | 0.665905 | 3 | 0 | 3 |
| PKHD1L1 | FAT4 | 0.665396 | 3 | 0 | 3 |
| PKHD1L1 | RCAN2 | 0.660956 | 3 | 0 | 3 |
| PKHD1L1 | FBLN1 | 0.653837 | 3 | 0 | 3 |
| PKHD1L1 | KCNMA1 | 0.650226 | 3 | 0 | 3 |
| PKHD1L1 | TMC8 | 0.650066 | 3 | 0 | 3 |
| PKHD1L1 | CLEC1A | 0.643271 | 4 | 0 | 3 |
| PKHD1L1 | RNF150 | 0.639711 | 3 | 0 | 3 |
| PKHD1L1 | CASQ2 | 0.630382 | 3 | 0 | 3 |
| PKHD1L1 | FILIP1 | 0.628581 | 4 | 0 | 3 |
| PKHD1L1 | SVEP1 | 0.625953 | 3 | 0 | 3 |
| PKHD1L1 | FMOD | 0.62561 | 3 | 0 | 3 |
| PKHD1L1 | NBEA | 0.624881 | 3 | 0 | 3 |
For details and further investigation, click here