| Full name: potassium channel tetramerization domain containing 12 | Alias Symbol: KIAA1778|PFET1 | ||
| Type: protein-coding gene | Cytoband: 13q22.3 | ||
| Entrez ID: 115207 | HGNC ID: HGNC:14678 | Ensembl Gene: ENSG00000178695 | OMIM ID: 610521 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD12 | 115207 | 212192_at | -0.8298 | 0.0631 | |
| GSE20347 | KCTD12 | 115207 | 212188_at | -0.4609 | 0.0459 | |
| GSE23400 | KCTD12 | 115207 | 212192_at | -0.2849 | 0.2337 | |
| GSE26886 | KCTD12 | 115207 | 212188_at | 0.1371 | 0.7989 | |
| GSE29001 | KCTD12 | 115207 | 212192_at | -0.2123 | 0.7906 | |
| GSE38129 | KCTD12 | 115207 | 212192_at | -0.7997 | 0.0135 | |
| GSE45670 | KCTD12 | 115207 | 212192_at | -1.5680 | 0.0000 | |
| GSE53622 | KCTD12 | 115207 | 70696 | -0.9828 | 0.0000 | |
| GSE53624 | KCTD12 | 115207 | 70696 | -0.6802 | 0.0000 | |
| GSE63941 | KCTD12 | 115207 | 212192_at | -4.1576 | 0.0089 | |
| GSE77861 | KCTD12 | 115207 | 212188_at | 0.7389 | 0.0665 | |
| GSE97050 | KCTD12 | 115207 | A_33_P3240507 | -0.9188 | 0.0867 | |
| SRP007169 | KCTD12 | 115207 | RNAseq | 1.0172 | 0.0271 | |
| SRP008496 | KCTD12 | 115207 | RNAseq | 0.4181 | 0.3281 | |
| SRP064894 | KCTD12 | 115207 | RNAseq | 0.0915 | 0.7898 | |
| SRP133303 | KCTD12 | 115207 | RNAseq | -0.1424 | 0.6371 | |
| SRP159526 | KCTD12 | 115207 | RNAseq | -0.4843 | 0.2506 | |
| SRP193095 | KCTD12 | 115207 | RNAseq | 0.1695 | 0.4623 | |
| SRP219564 | KCTD12 | 115207 | RNAseq | -0.3770 | 0.5791 | |
| TCGA | KCTD12 | 115207 | RNAseq | -0.2476 | 0.0023 |
Upregulated datasets: 1; Downregulated datasets: 2.
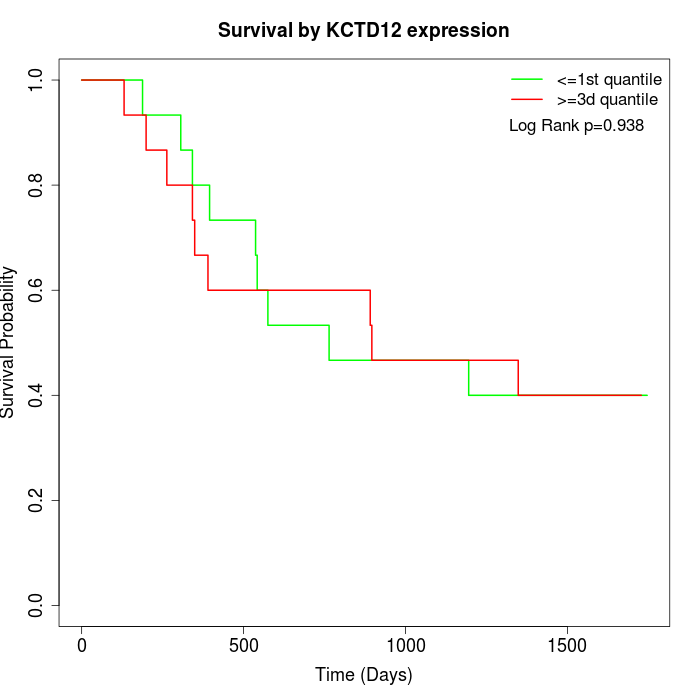
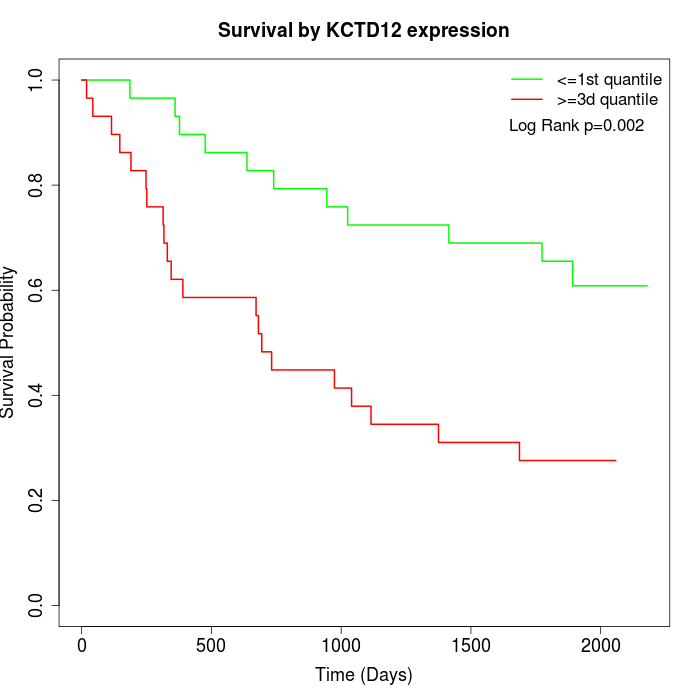
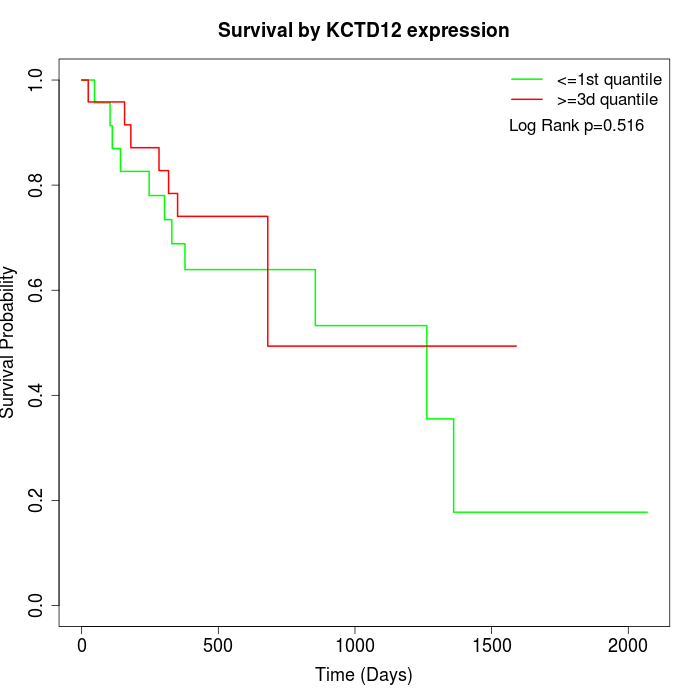
Survival by KCTD12 expression:
Note: Click image to view full size file.
Copy number change of KCTD12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD12 | 115207 | 3 | 11 | 16 | |
| GSE20123 | KCTD12 | 115207 | 3 | 10 | 17 | |
| GSE43470 | KCTD12 | 115207 | 4 | 11 | 28 | |
| GSE46452 | KCTD12 | 115207 | 0 | 32 | 27 | |
| GSE47630 | KCTD12 | 115207 | 3 | 27 | 10 | |
| GSE54993 | KCTD12 | 115207 | 11 | 3 | 56 | |
| GSE54994 | KCTD12 | 115207 | 3 | 15 | 35 | |
| GSE60625 | KCTD12 | 115207 | 0 | 3 | 8 | |
| GSE74703 | KCTD12 | 115207 | 3 | 9 | 24 | |
| GSE74704 | KCTD12 | 115207 | 1 | 9 | 10 | |
| TCGA | KCTD12 | 115207 | 12 | 33 | 51 |
Total number of gains: 43; Total number of losses: 163; Total Number of normals: 282.
Somatic mutations of KCTD12:
Generating mutation plots.
Highly correlated genes for KCTD12:
Showing top 20/948 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD12 | LRFN5 | 0.78348 | 4 | 0 | 4 |
| KCTD12 | TNFSF12 | 0.773391 | 4 | 0 | 4 |
| KCTD12 | GNPAT | 0.76967 | 3 | 0 | 3 |
| KCTD12 | ZEB1 | 0.769034 | 3 | 0 | 3 |
| KCTD12 | SORCS1 | 0.767534 | 4 | 0 | 4 |
| KCTD12 | SPARCL1 | 0.765474 | 9 | 0 | 9 |
| KCTD12 | CLEC3B | 0.763594 | 3 | 0 | 3 |
| KCTD12 | CYP21A2 | 0.762656 | 3 | 0 | 3 |
| KCTD12 | MSRB3 | 0.756857 | 7 | 0 | 7 |
| KCTD12 | AP2B1 | 0.756733 | 3 | 0 | 3 |
| KCTD12 | GNG2 | 0.756477 | 5 | 0 | 5 |
| KCTD12 | AGTR1 | 0.750031 | 8 | 0 | 8 |
| KCTD12 | CILP | 0.746068 | 8 | 0 | 7 |
| KCTD12 | LINC01279 | 0.745437 | 4 | 0 | 4 |
| KCTD12 | COLEC12 | 0.738699 | 9 | 0 | 9 |
| KCTD12 | TMEM47 | 0.738614 | 12 | 0 | 11 |
| KCTD12 | ZFAND5 | 0.738111 | 3 | 0 | 3 |
| KCTD12 | FILIP1 | 0.736133 | 5 | 0 | 5 |
| KCTD12 | ZCCHC24 | 0.736062 | 10 | 0 | 9 |
| KCTD12 | CFL2 | 0.735001 | 5 | 0 | 5 |
For details and further investigation, click here