| Full name: pleckstrin homology and RhoGEF domain containing G3 | Alias Symbol: ARHGEF43 | ||
| Type: protein-coding gene | Cytoband: 14q23.3 | ||
| Entrez ID: 26030 | HGNC ID: HGNC:20364 | Ensembl Gene: ENSG00000126822 | OMIM ID: 617940 |
| Drug and gene relationship at DGIdb | |||
Expression of PLEKHG3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLEKHG3 | 26030 | 212823_s_at | 0.2038 | 0.6627 | |
| GSE20347 | PLEKHG3 | 26030 | 212823_s_at | 0.0682 | 0.6037 | |
| GSE23400 | PLEKHG3 | 26030 | 212823_s_at | 0.1270 | 0.0179 | |
| GSE26886 | PLEKHG3 | 26030 | 212823_s_at | 0.4547 | 0.0038 | |
| GSE29001 | PLEKHG3 | 26030 | 212823_s_at | -0.0656 | 0.7840 | |
| GSE38129 | PLEKHG3 | 26030 | 212823_s_at | 0.1459 | 0.3067 | |
| GSE45670 | PLEKHG3 | 26030 | 212823_s_at | 0.2353 | 0.0809 | |
| GSE53622 | PLEKHG3 | 26030 | 38302 | -0.1573 | 0.1313 | |
| GSE53624 | PLEKHG3 | 26030 | 38302 | 0.0247 | 0.7353 | |
| GSE63941 | PLEKHG3 | 26030 | 212823_s_at | 1.2354 | 0.0005 | |
| GSE77861 | PLEKHG3 | 26030 | 212823_s_at | 0.1577 | 0.5422 | |
| GSE97050 | PLEKHG3 | 26030 | A_23_P76901 | -0.0083 | 0.9762 | |
| SRP007169 | PLEKHG3 | 26030 | RNAseq | 0.7989 | 0.0453 | |
| SRP008496 | PLEKHG3 | 26030 | RNAseq | 0.8338 | 0.0004 | |
| SRP064894 | PLEKHG3 | 26030 | RNAseq | 0.3046 | 0.2618 | |
| SRP133303 | PLEKHG3 | 26030 | RNAseq | 0.3022 | 0.3318 | |
| SRP159526 | PLEKHG3 | 26030 | RNAseq | -0.0546 | 0.9137 | |
| SRP193095 | PLEKHG3 | 26030 | RNAseq | 0.5446 | 0.0009 | |
| SRP219564 | PLEKHG3 | 26030 | RNAseq | 0.2738 | 0.6537 | |
| TCGA | PLEKHG3 | 26030 | RNAseq | 0.1506 | 0.0383 |
Upregulated datasets: 1; Downregulated datasets: 0.
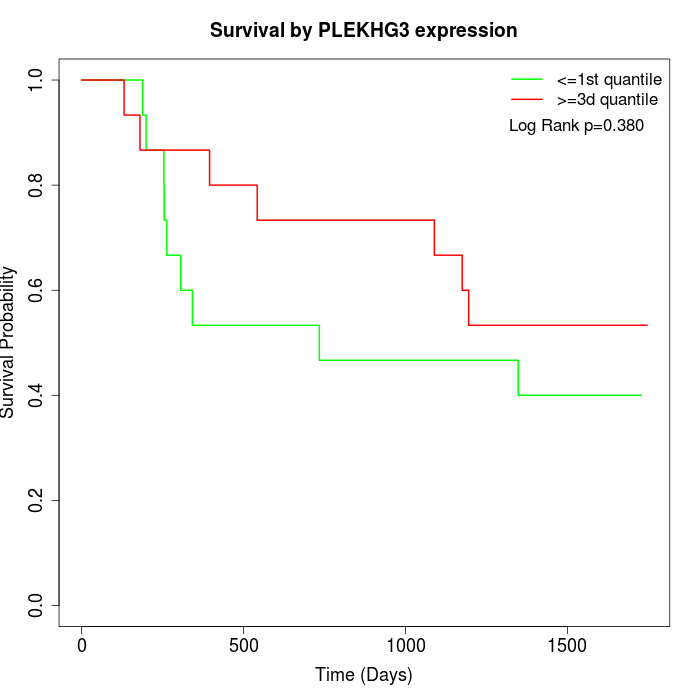
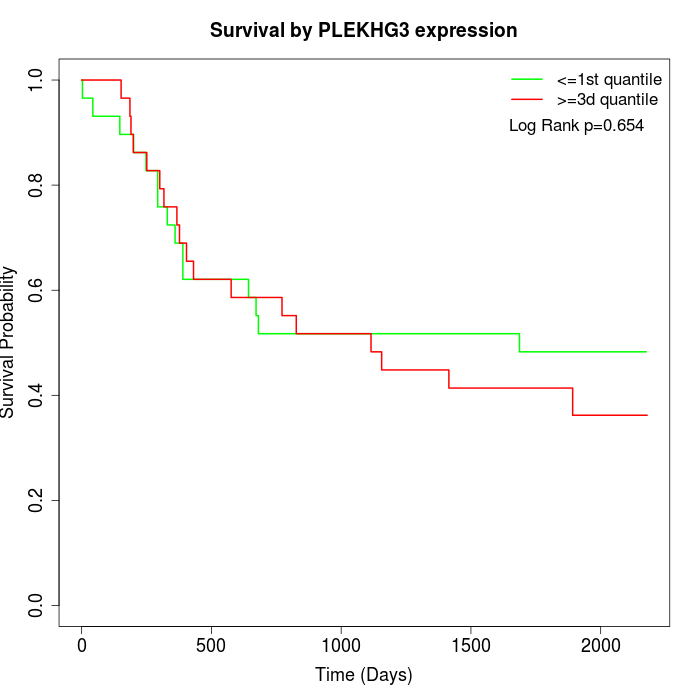
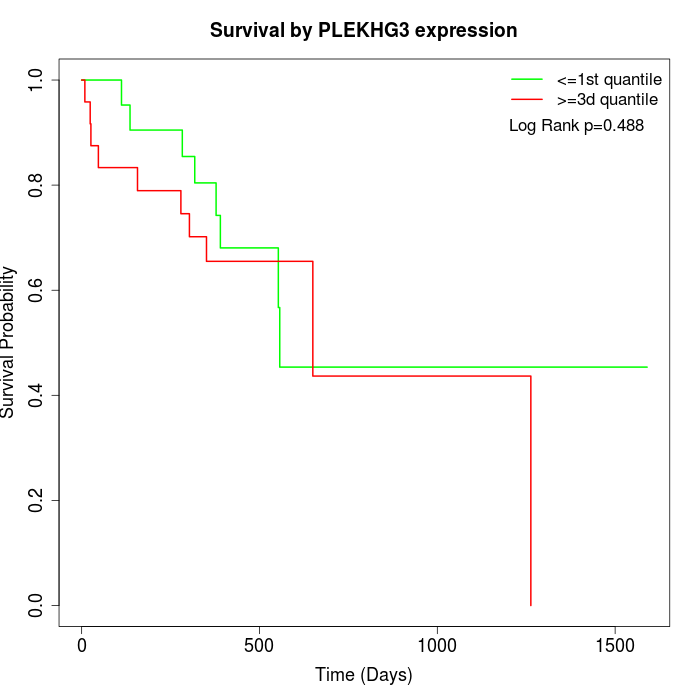
Survival by PLEKHG3 expression:
Note: Click image to view full size file.
Copy number change of PLEKHG3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLEKHG3 | 26030 | 7 | 3 | 20 | |
| GSE20123 | PLEKHG3 | 26030 | 7 | 3 | 20 | |
| GSE43470 | PLEKHG3 | 26030 | 8 | 2 | 33 | |
| GSE46452 | PLEKHG3 | 26030 | 16 | 3 | 40 | |
| GSE47630 | PLEKHG3 | 26030 | 11 | 9 | 20 | |
| GSE54993 | PLEKHG3 | 26030 | 3 | 9 | 58 | |
| GSE54994 | PLEKHG3 | 26030 | 18 | 5 | 30 | |
| GSE60625 | PLEKHG3 | 26030 | 0 | 2 | 9 | |
| GSE74703 | PLEKHG3 | 26030 | 7 | 2 | 27 | |
| GSE74704 | PLEKHG3 | 26030 | 3 | 2 | 15 | |
| TCGA | PLEKHG3 | 26030 | 32 | 14 | 50 |
Total number of gains: 112; Total number of losses: 54; Total Number of normals: 322.
Somatic mutations of PLEKHG3:
Generating mutation plots.
Highly correlated genes for PLEKHG3:
Showing top 20/414 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLEKHG3 | CD22 | 0.733309 | 3 | 0 | 3 |
| PLEKHG3 | CCNA1 | 0.728138 | 3 | 0 | 3 |
| PLEKHG3 | LSG1 | 0.688066 | 3 | 0 | 3 |
| PLEKHG3 | DMAP1 | 0.683439 | 3 | 0 | 3 |
| PLEKHG3 | NUDT14 | 0.672882 | 4 | 0 | 3 |
| PLEKHG3 | INO80B | 0.656568 | 6 | 0 | 5 |
| PLEKHG3 | TIGD1 | 0.653932 | 4 | 0 | 4 |
| PLEKHG3 | FAM110C | 0.649387 | 3 | 0 | 3 |
| PLEKHG3 | DND1 | 0.648373 | 3 | 0 | 3 |
| PLEKHG3 | RPL8 | 0.647768 | 5 | 0 | 3 |
| PLEKHG3 | C6orf163 | 0.640695 | 3 | 0 | 3 |
| PLEKHG3 | NCOA6 | 0.639171 | 4 | 0 | 3 |
| PLEKHG3 | OR12D2 | 0.635137 | 3 | 0 | 3 |
| PLEKHG3 | GGA2 | 0.63429 | 4 | 0 | 3 |
| PLEKHG3 | DVL2 | 0.634092 | 3 | 0 | 3 |
| PLEKHG3 | PIGL | 0.632252 | 7 | 0 | 6 |
| PLEKHG3 | IRF2BPL | 0.631802 | 3 | 0 | 3 |
| PLEKHG3 | NEURL1B | 0.631309 | 3 | 0 | 3 |
| PLEKHG3 | MXD3 | 0.628283 | 3 | 0 | 3 |
| PLEKHG3 | PKM | 0.627813 | 5 | 0 | 4 |
For details and further investigation, click here