| Full name: protein phosphatase, Mg2+/Mn2+ dependent 1B | Alias Symbol: PPC2BETAX|PP2CB|PP2CBETA | ||
| Type: protein-coding gene | Cytoband: 2p21 | ||
| Entrez ID: 5495 | HGNC ID: HGNC:9276 | Ensembl Gene: ENSG00000138032 | OMIM ID: 603770 |
| Drug and gene relationship at DGIdb | |||
PPM1B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway |
Expression of PPM1B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPM1B | 5495 | 209296_at | 0.0923 | 0.8093 | |
| GSE20347 | PPM1B | 5495 | 209296_at | 0.0554 | 0.7748 | |
| GSE23400 | PPM1B | 5495 | 209296_at | 0.0098 | 0.9090 | |
| GSE26886 | PPM1B | 5495 | 209296_at | -1.1677 | 0.0000 | |
| GSE29001 | PPM1B | 5495 | 209296_at | 0.2438 | 0.2651 | |
| GSE38129 | PPM1B | 5495 | 209296_at | -0.0918 | 0.5188 | |
| GSE45670 | PPM1B | 5495 | 209296_at | -0.0473 | 0.8106 | |
| GSE53622 | PPM1B | 5495 | 48781 | -0.1070 | 0.0856 | |
| GSE53624 | PPM1B | 5495 | 48781 | -0.2400 | 0.0004 | |
| GSE63941 | PPM1B | 5495 | 209296_at | -0.1667 | 0.7064 | |
| GSE77861 | PPM1B | 5495 | 209296_at | 0.3882 | 0.1919 | |
| GSE97050 | PPM1B | 5495 | A_33_P3296352 | -0.2316 | 0.3948 | |
| SRP007169 | PPM1B | 5495 | RNAseq | 0.7304 | 0.0859 | |
| SRP008496 | PPM1B | 5495 | RNAseq | 0.7148 | 0.0094 | |
| SRP064894 | PPM1B | 5495 | RNAseq | -0.3551 | 0.0938 | |
| SRP133303 | PPM1B | 5495 | RNAseq | 0.1551 | 0.1920 | |
| SRP159526 | PPM1B | 5495 | RNAseq | 0.0933 | 0.6757 | |
| SRP193095 | PPM1B | 5495 | RNAseq | -0.0188 | 0.8329 | |
| SRP219564 | PPM1B | 5495 | RNAseq | -0.2596 | 0.3057 | |
| TCGA | PPM1B | 5495 | RNAseq | -0.0827 | 0.0783 |
Upregulated datasets: 0; Downregulated datasets: 1.
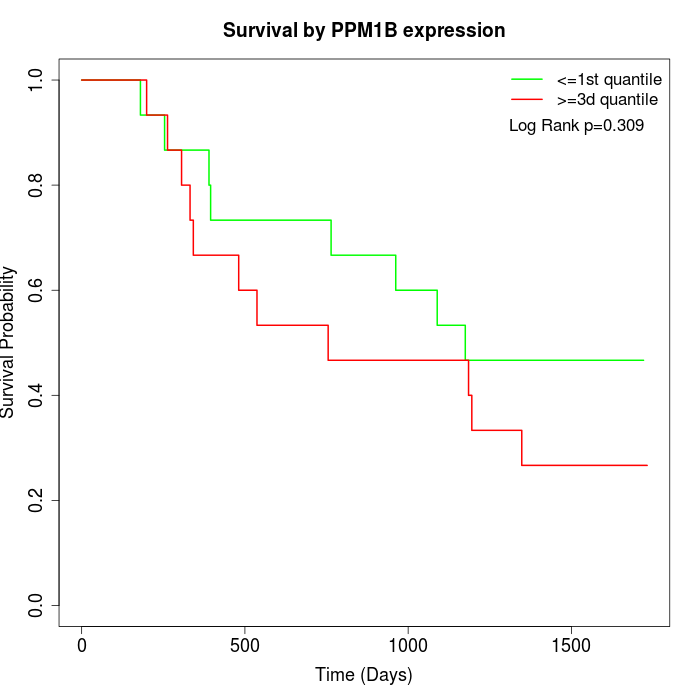
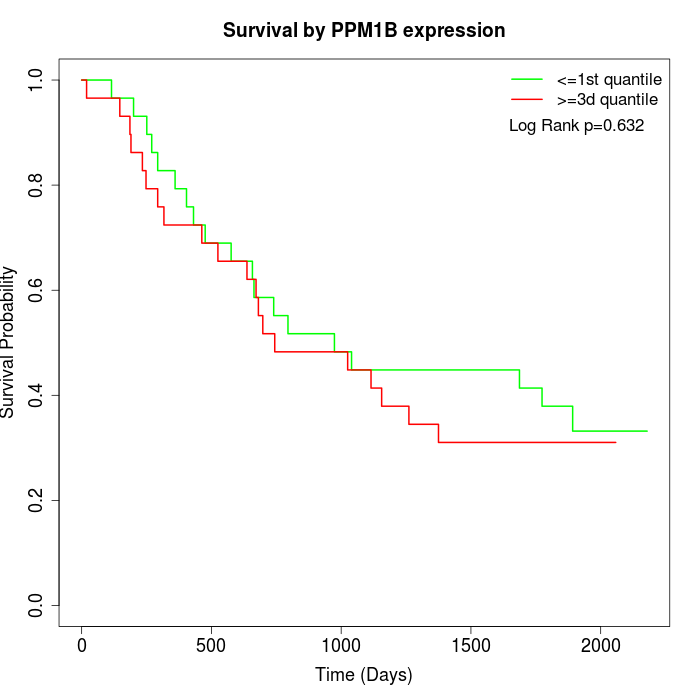
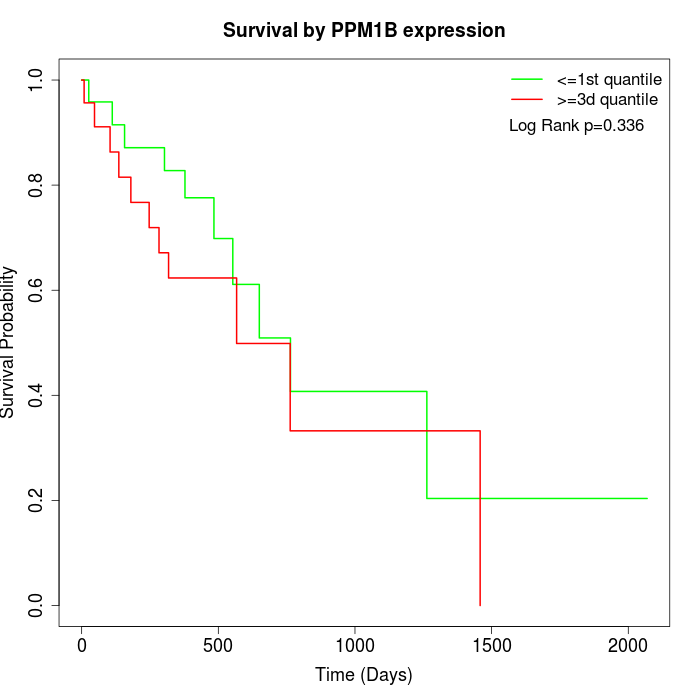
Survival by PPM1B expression:
Note: Click image to view full size file.
Copy number change of PPM1B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPM1B | 5495 | 10 | 1 | 19 | |
| GSE20123 | PPM1B | 5495 | 9 | 1 | 20 | |
| GSE43470 | PPM1B | 5495 | 4 | 0 | 39 | |
| GSE46452 | PPM1B | 5495 | 1 | 4 | 54 | |
| GSE47630 | PPM1B | 5495 | 8 | 0 | 32 | |
| GSE54993 | PPM1B | 5495 | 0 | 5 | 65 | |
| GSE54994 | PPM1B | 5495 | 10 | 0 | 43 | |
| GSE60625 | PPM1B | 5495 | 0 | 3 | 8 | |
| GSE74703 | PPM1B | 5495 | 4 | 0 | 32 | |
| GSE74704 | PPM1B | 5495 | 9 | 1 | 10 | |
| TCGA | PPM1B | 5495 | 35 | 2 | 59 |
Total number of gains: 90; Total number of losses: 17; Total Number of normals: 381.
Somatic mutations of PPM1B:
Generating mutation plots.
Highly correlated genes for PPM1B:
Showing top 20/260 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPM1B | STX17 | 0.811213 | 3 | 0 | 3 |
| PPM1B | EXOC2 | 0.808701 | 3 | 0 | 3 |
| PPM1B | DCP1A | 0.791188 | 3 | 0 | 3 |
| PPM1B | TP53BP2 | 0.782625 | 3 | 0 | 3 |
| PPM1B | MTMR9 | 0.766098 | 3 | 0 | 3 |
| PPM1B | CWC25 | 0.752859 | 3 | 0 | 3 |
| PPM1B | ETFA | 0.752097 | 3 | 0 | 3 |
| PPM1B | METTL14 | 0.73506 | 3 | 0 | 3 |
| PPM1B | HNRNPF | 0.727754 | 3 | 0 | 3 |
| PPM1B | FH | 0.722586 | 3 | 0 | 3 |
| PPM1B | RASA2 | 0.713129 | 3 | 0 | 3 |
| PPM1B | PLEKHF2 | 0.710627 | 3 | 0 | 3 |
| PPM1B | AKAP17A | 0.708181 | 3 | 0 | 3 |
| PPM1B | HBS1L | 0.707774 | 5 | 0 | 5 |
| PPM1B | TRIM59 | 0.701776 | 3 | 0 | 3 |
| PPM1B | CNST | 0.698036 | 4 | 0 | 3 |
| PPM1B | NEDD4L | 0.697819 | 3 | 0 | 3 |
| PPM1B | WLS | 0.697335 | 3 | 0 | 3 |
| PPM1B | MSI2 | 0.690959 | 3 | 0 | 3 |
| PPM1B | SNX17 | 0.683083 | 5 | 0 | 5 |
For details and further investigation, click here