| Full name: RAB25, member RAS oncogene family | Alias Symbol: CATX-8 | ||
| Type: protein-coding gene | Cytoband: 1q22 | ||
| Entrez ID: 57111 | HGNC ID: HGNC:18238 | Ensembl Gene: ENSG00000132698 | OMIM ID: 612942 |
| Drug and gene relationship at DGIdb | |||
Expression of RAB25:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAB25 | 57111 | 218186_at | -0.8836 | 0.2715 | |
| GSE20347 | RAB25 | 57111 | 218186_at | -2.2066 | 0.0000 | |
| GSE23400 | RAB25 | 57111 | 218186_at | -1.6298 | 0.0000 | |
| GSE26886 | RAB25 | 57111 | 218186_at | -2.6763 | 0.0000 | |
| GSE29001 | RAB25 | 57111 | 218186_at | -2.4562 | 0.0003 | |
| GSE38129 | RAB25 | 57111 | 218186_at | -1.0257 | 0.0763 | |
| GSE45670 | RAB25 | 57111 | 218186_at | -0.6990 | 0.0230 | |
| GSE53622 | RAB25 | 57111 | 74303 | -1.6735 | 0.0000 | |
| GSE53624 | RAB25 | 57111 | 74303 | -2.2648 | 0.0000 | |
| GSE63941 | RAB25 | 57111 | 218186_at | 5.6161 | 0.0004 | |
| GSE77861 | RAB25 | 57111 | 218186_at | -1.2736 | 0.0011 | |
| GSE97050 | RAB25 | 57111 | A_23_P115091 | -0.3139 | 0.7690 | |
| SRP007169 | RAB25 | 57111 | RNAseq | -3.4376 | 0.0000 | |
| SRP008496 | RAB25 | 57111 | RNAseq | -3.4953 | 0.0000 | |
| SRP064894 | RAB25 | 57111 | RNAseq | -2.1038 | 0.0000 | |
| SRP133303 | RAB25 | 57111 | RNAseq | -2.2886 | 0.0000 | |
| SRP159526 | RAB25 | 57111 | RNAseq | -2.0678 | 0.0000 | |
| SRP193095 | RAB25 | 57111 | RNAseq | -2.1534 | 0.0000 | |
| SRP219564 | RAB25 | 57111 | RNAseq | -2.0440 | 0.1329 | |
| TCGA | RAB25 | 57111 | RNAseq | 0.1243 | 0.5012 |
Upregulated datasets: 1; Downregulated datasets: 13.
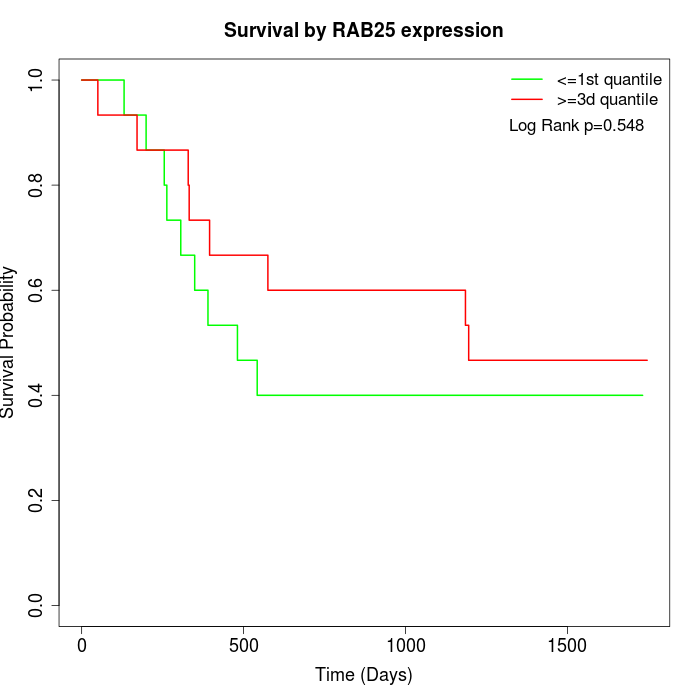
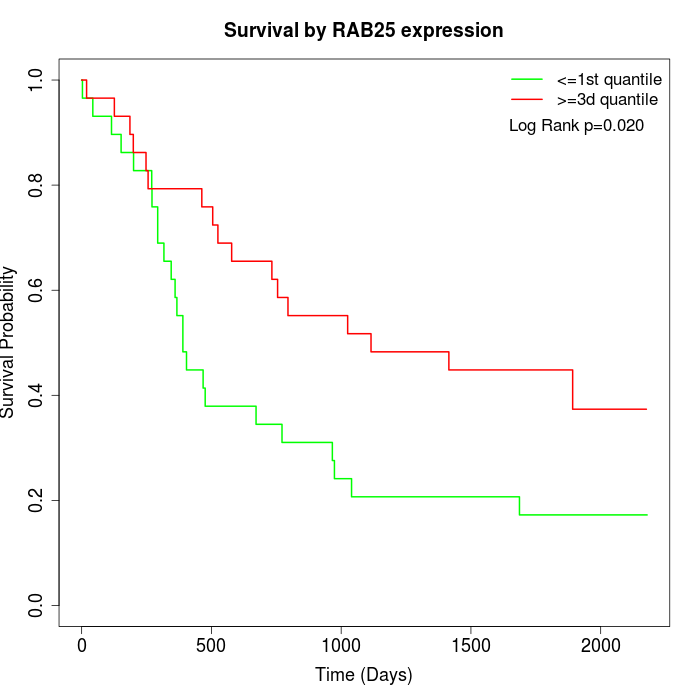
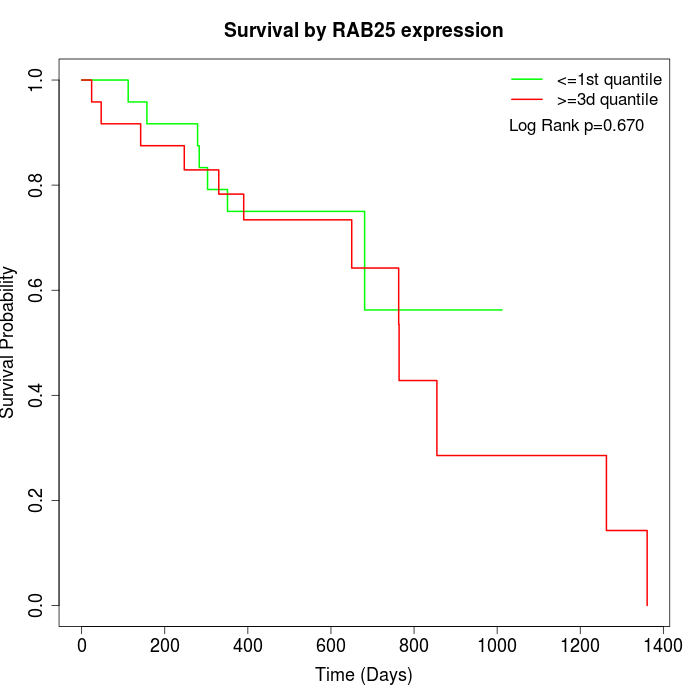
Survival by RAB25 expression:
Note: Click image to view full size file.
Copy number change of RAB25:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAB25 | 57111 | 14 | 0 | 16 | |
| GSE20123 | RAB25 | 57111 | 13 | 0 | 17 | |
| GSE43470 | RAB25 | 57111 | 7 | 2 | 34 | |
| GSE46452 | RAB25 | 57111 | 2 | 1 | 56 | |
| GSE47630 | RAB25 | 57111 | 14 | 0 | 26 | |
| GSE54993 | RAB25 | 57111 | 0 | 5 | 65 | |
| GSE54994 | RAB25 | 57111 | 16 | 0 | 37 | |
| GSE60625 | RAB25 | 57111 | 0 | 0 | 11 | |
| GSE74703 | RAB25 | 57111 | 7 | 2 | 27 | |
| GSE74704 | RAB25 | 57111 | 6 | 0 | 14 | |
| TCGA | RAB25 | 57111 | 38 | 2 | 56 |
Total number of gains: 117; Total number of losses: 12; Total Number of normals: 359.
Somatic mutations of RAB25:
Generating mutation plots.
Highly correlated genes for RAB25:
Showing top 20/1631 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAB25 | NIPAL1 | 0.862083 | 3 | 0 | 3 |
| RAB25 | ULK3 | 0.837417 | 7 | 0 | 7 |
| RAB25 | DEGS2 | 0.837072 | 6 | 0 | 6 |
| RAB25 | PLEKHA7 | 0.833551 | 8 | 0 | 8 |
| RAB25 | RASSF5 | 0.832899 | 7 | 0 | 7 |
| RAB25 | SFTA2 | 0.828882 | 4 | 0 | 4 |
| RAB25 | EPHA1 | 0.824588 | 12 | 0 | 12 |
| RAB25 | EVPL | 0.824223 | 12 | 0 | 11 |
| RAB25 | VSIG10L | 0.822845 | 6 | 0 | 6 |
| RAB25 | MPZL3 | 0.822453 | 8 | 0 | 8 |
| RAB25 | CLIC3 | 0.815094 | 10 | 0 | 10 |
| RAB25 | NT5C2 | 0.814082 | 11 | 0 | 11 |
| RAB25 | ARHGAP27 | 0.813788 | 9 | 0 | 9 |
| RAB25 | BARX2 | 0.810308 | 10 | 0 | 10 |
| RAB25 | TOM1 | 0.810267 | 11 | 0 | 11 |
| RAB25 | EPGN | 0.80765 | 3 | 0 | 3 |
| RAB25 | SCNN1B | 0.807568 | 11 | 0 | 10 |
| RAB25 | TIAM1 | 0.806794 | 12 | 0 | 12 |
| RAB25 | C6orf132 | 0.805818 | 8 | 0 | 7 |
| RAB25 | CLDN7 | 0.803552 | 13 | 0 | 13 |
For details and further investigation, click here