| Full name: RAB3C, member RAS oncogene family | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5q11.2 | ||
| Entrez ID: 115827 | HGNC ID: HGNC:30269 | Ensembl Gene: ENSG00000152932 | OMIM ID: 612829 |
| Drug and gene relationship at DGIdb | |||
Expression of RAB3C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAB3C | 115827 | 229300_at | -0.2617 | 0.4550 | |
| GSE26886 | RAB3C | 115827 | 229300_at | -0.0239 | 0.8823 | |
| GSE45670 | RAB3C | 115827 | 229300_at | 0.0974 | 0.3919 | |
| GSE53622 | RAB3C | 115827 | 4406 | -2.1086 | 0.0000 | |
| GSE53624 | RAB3C | 115827 | 91184 | -0.3867 | 0.0000 | |
| GSE63941 | RAB3C | 115827 | 229300_at | 0.0493 | 0.7256 | |
| GSE77861 | RAB3C | 115827 | 229300_at | -0.0923 | 0.4369 | |
| SRP133303 | RAB3C | 115827 | RNAseq | -1.3047 | 0.0083 | |
| TCGA | RAB3C | 115827 | RNAseq | -3.2740 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
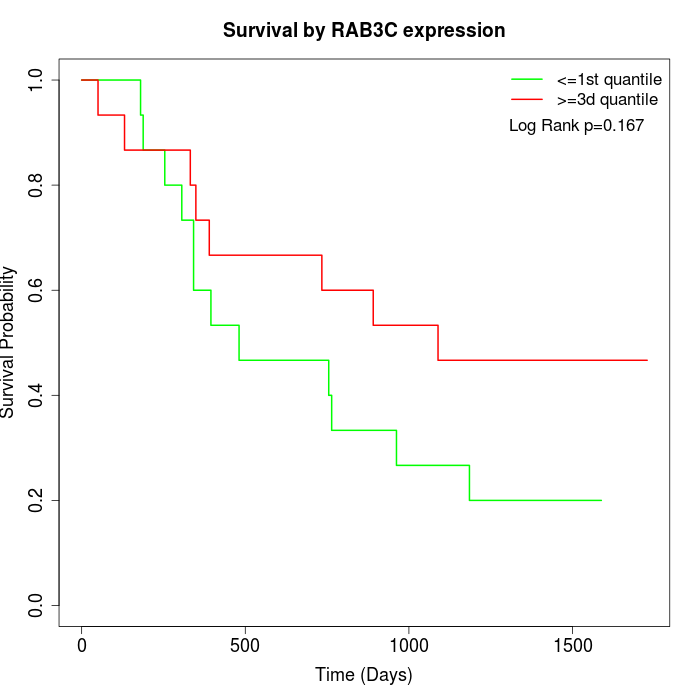
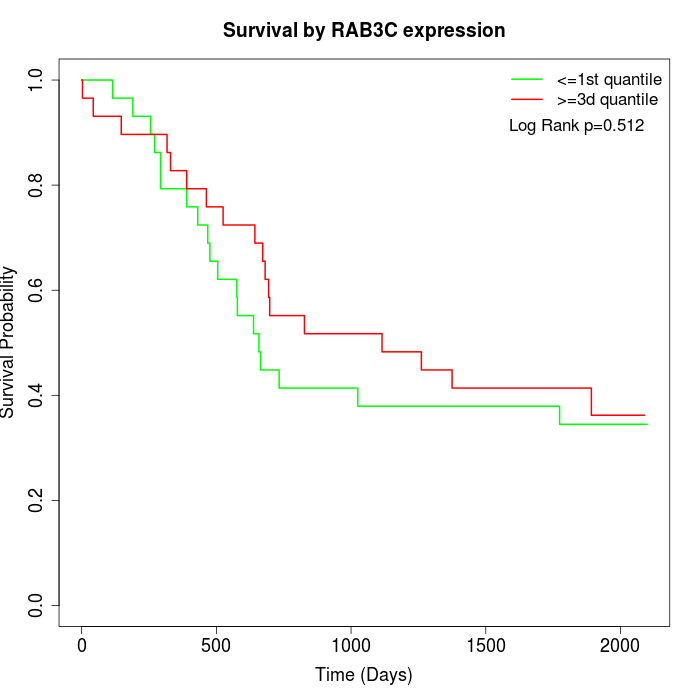
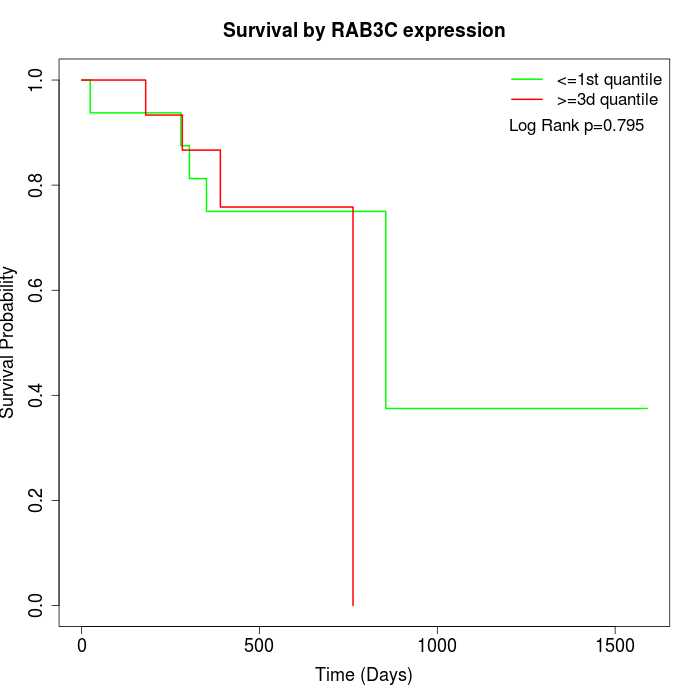
Survival by RAB3C expression:
Note: Click image to view full size file.
Copy number change of RAB3C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAB3C | 115827 | 0 | 13 | 17 | |
| GSE20123 | RAB3C | 115827 | 0 | 13 | 17 | |
| GSE43470 | RAB3C | 115827 | 1 | 12 | 30 | |
| GSE46452 | RAB3C | 115827 | 0 | 28 | 31 | |
| GSE47630 | RAB3C | 115827 | 2 | 16 | 22 | |
| GSE54993 | RAB3C | 115827 | 8 | 1 | 61 | |
| GSE54994 | RAB3C | 115827 | 2 | 19 | 32 | |
| GSE60625 | RAB3C | 115827 | 0 | 0 | 11 | |
| GSE74703 | RAB3C | 115827 | 1 | 9 | 26 | |
| GSE74704 | RAB3C | 115827 | 0 | 6 | 14 | |
| TCGA | RAB3C | 115827 | 7 | 47 | 42 |
Total number of gains: 21; Total number of losses: 164; Total Number of normals: 303.
Somatic mutations of RAB3C:
Generating mutation plots.
Highly correlated genes for RAB3C:
Showing top 20/54 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAB3C | ATOH8 | 0.742605 | 3 | 0 | 3 |
| RAB3C | TAGLN | 0.708002 | 3 | 0 | 3 |
| RAB3C | BMX | 0.68605 | 3 | 0 | 3 |
| RAB3C | ADAMTS8 | 0.682228 | 3 | 0 | 3 |
| RAB3C | SOD3 | 0.67763 | 3 | 0 | 3 |
| RAB3C | PGM5 | 0.675513 | 3 | 0 | 3 |
| RAB3C | AOX1 | 0.675307 | 3 | 0 | 3 |
| RAB3C | LMOD1 | 0.675302 | 3 | 0 | 3 |
| RAB3C | PDZRN4 | 0.661712 | 3 | 0 | 3 |
| RAB3C | FAM124A | 0.661076 | 3 | 0 | 3 |
| RAB3C | RAMP2 | 0.647336 | 4 | 0 | 4 |
| RAB3C | DACT3 | 0.646318 | 3 | 0 | 3 |
| RAB3C | OXCT1 | 0.640365 | 3 | 0 | 3 |
| RAB3C | RPS6KA2 | 0.634028 | 3 | 0 | 3 |
| RAB3C | ARHGAP6 | 0.627604 | 4 | 0 | 3 |
| RAB3C | SORBS2 | 0.62486 | 3 | 0 | 3 |
| RAB3C | OSBPL1A | 0.618548 | 3 | 0 | 3 |
| RAB3C | ADAMTS9-AS2 | 0.615075 | 3 | 0 | 3 |
| RAB3C | PARD3B | 0.611189 | 3 | 0 | 3 |
| RAB3C | GNPDA2 | 0.609353 | 3 | 0 | 3 |
For details and further investigation, click here