| Full name: Rab geranylgeranyltransferase subunit alpha | Alias Symbol: PTAR3 | ||
| Type: protein-coding gene | Cytoband: 14q11.2 | ||
| Entrez ID: 5875 | HGNC ID: HGNC:9795 | Ensembl Gene: ENSG00000100949 | OMIM ID: 601905 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RABGGTA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RABGGTA | 5875 | 203573_s_at | -0.6611 | 0.4968 | |
| GSE20347 | RABGGTA | 5875 | 203573_s_at | -1.2292 | 0.0000 | |
| GSE23400 | RABGGTA | 5875 | 203573_s_at | -0.7546 | 0.0000 | |
| GSE26886 | RABGGTA | 5875 | 203573_s_at | -1.3554 | 0.0000 | |
| GSE29001 | RABGGTA | 5875 | 203573_s_at | -1.1584 | 0.0003 | |
| GSE38129 | RABGGTA | 5875 | 203573_s_at | -0.8147 | 0.0021 | |
| GSE45670 | RABGGTA | 5875 | 203573_s_at | 0.0561 | 0.8220 | |
| GSE53622 | RABGGTA | 5875 | 173831 | -1.1742 | 0.0000 | |
| GSE53624 | RABGGTA | 5875 | 66837 | -1.3231 | 0.0000 | |
| GSE63941 | RABGGTA | 5875 | 203573_s_at | 0.7912 | 0.1084 | |
| GSE77861 | RABGGTA | 5875 | 203573_s_at | -0.5294 | 0.0601 | |
| GSE97050 | RABGGTA | 5875 | A_23_P54223 | 0.1373 | 0.7054 | |
| SRP007169 | RABGGTA | 5875 | RNAseq | -2.1480 | 0.0000 | |
| SRP008496 | RABGGTA | 5875 | RNAseq | -2.6090 | 0.0000 | |
| SRP064894 | RABGGTA | 5875 | RNAseq | -0.6427 | 0.0051 | |
| SRP133303 | RABGGTA | 5875 | RNAseq | -0.9204 | 0.0001 | |
| SRP159526 | RABGGTA | 5875 | RNAseq | -1.3013 | 0.0000 | |
| SRP193095 | RABGGTA | 5875 | RNAseq | -0.9785 | 0.0000 | |
| SRP219564 | RABGGTA | 5875 | RNAseq | -1.0025 | 0.1072 | |
| TCGA | RABGGTA | 5875 | RNAseq | 0.0528 | 0.4761 |
Upregulated datasets: 0; Downregulated datasets: 8.
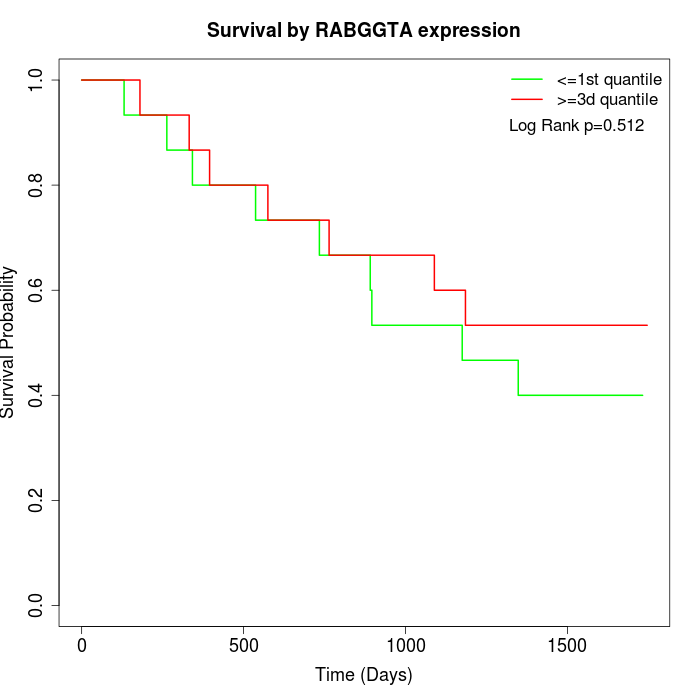
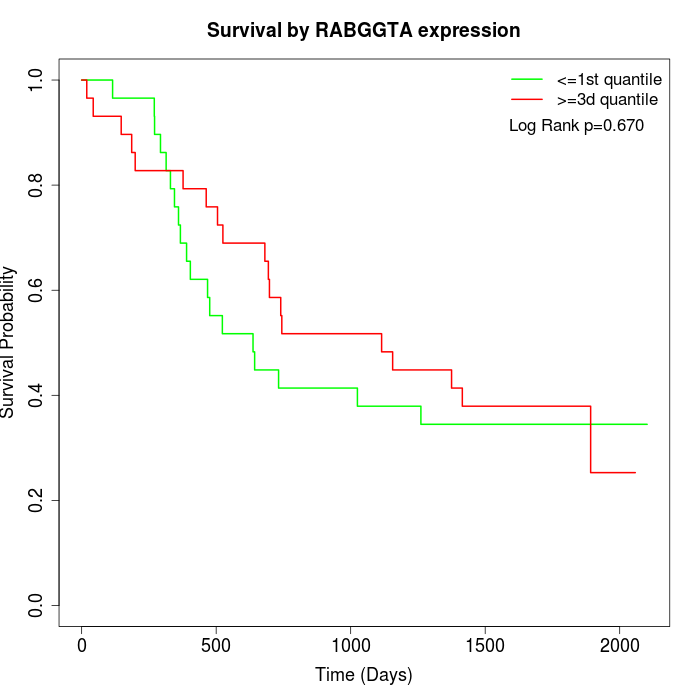
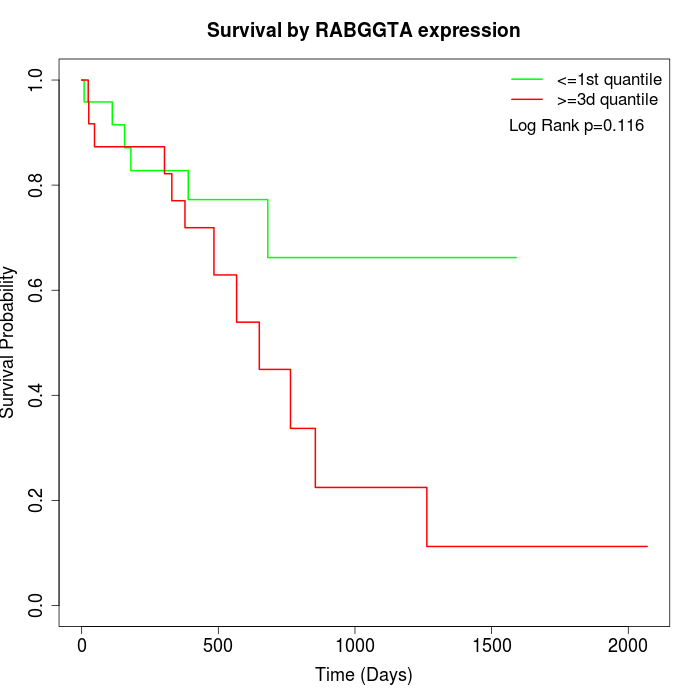
Survival by RABGGTA expression:
Note: Click image to view full size file.
Copy number change of RABGGTA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RABGGTA | 5875 | 8 | 3 | 19 | |
| GSE20123 | RABGGTA | 5875 | 8 | 3 | 19 | |
| GSE43470 | RABGGTA | 5875 | 8 | 2 | 33 | |
| GSE46452 | RABGGTA | 5875 | 16 | 2 | 41 | |
| GSE47630 | RABGGTA | 5875 | 11 | 10 | 19 | |
| GSE54993 | RABGGTA | 5875 | 3 | 11 | 56 | |
| GSE54994 | RABGGTA | 5875 | 18 | 5 | 30 | |
| GSE60625 | RABGGTA | 5875 | 0 | 2 | 9 | |
| GSE74703 | RABGGTA | 5875 | 7 | 2 | 27 | |
| GSE74704 | RABGGTA | 5875 | 3 | 2 | 15 | |
| TCGA | RABGGTA | 5875 | 27 | 12 | 57 |
Total number of gains: 109; Total number of losses: 54; Total Number of normals: 325.
Somatic mutations of RABGGTA:
Generating mutation plots.
Highly correlated genes for RABGGTA:
Showing top 20/1578 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RABGGTA | CHCHD1 | 0.805152 | 3 | 0 | 3 |
| RABGGTA | TMEM167A | 0.802505 | 3 | 0 | 3 |
| RABGGTA | TGM1 | 0.799796 | 13 | 0 | 12 |
| RABGGTA | NBEAL2 | 0.797768 | 10 | 0 | 10 |
| RABGGTA | ARPC1A | 0.797224 | 3 | 0 | 3 |
| RABGGTA | TOM1 | 0.796722 | 12 | 0 | 11 |
| RABGGTA | DDI2 | 0.793538 | 3 | 0 | 3 |
| RABGGTA | BLOC1S2 | 0.790849 | 4 | 0 | 4 |
| RABGGTA | ALS2CL | 0.790503 | 10 | 0 | 10 |
| RABGGTA | ASCC2 | 0.787522 | 11 | 0 | 11 |
| RABGGTA | SULT2B1 | 0.786404 | 9 | 0 | 9 |
| RABGGTA | CCDC7 | 0.785318 | 3 | 0 | 3 |
| RABGGTA | DHRS1 | 0.776088 | 11 | 0 | 10 |
| RABGGTA | CRABP2 | 0.773675 | 10 | 0 | 10 |
| RABGGTA | TMEM40 | 0.766161 | 12 | 0 | 11 |
| RABGGTA | NLRX1 | 0.765522 | 11 | 0 | 10 |
| RABGGTA | ZNF185 | 0.765351 | 11 | 0 | 11 |
| RABGGTA | LYPD3 | 0.765237 | 11 | 0 | 11 |
| RABGGTA | RSPH3 | 0.764978 | 3 | 0 | 3 |
| RABGGTA | ECM1 | 0.763996 | 10 | 0 | 10 |
For details and further investigation, click here