| Full name: Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | Alias Symbol: KIAA1681 | ||
| Type: protein-coding gene | Cytoband: 2q33.2 | ||
| Entrez ID: 65059 | HGNC ID: HGNC:14436 | Ensembl Gene: ENSG00000173166 | OMIM ID: 609035 |
| Drug and gene relationship at DGIdb | |||
Expression of RAPH1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAPH1 | 65059 | 225188_at | 0.2763 | 0.6871 | |
| GSE26886 | RAPH1 | 65059 | 225188_at | 0.8513 | 0.0023 | |
| GSE45670 | RAPH1 | 65059 | 225188_at | -0.1008 | 0.6957 | |
| GSE53622 | RAPH1 | 65059 | 75665 | 0.0276 | 0.7436 | |
| GSE53624 | RAPH1 | 65059 | 75665 | 0.0268 | 0.6860 | |
| GSE63941 | RAPH1 | 65059 | 225188_at | -2.0239 | 0.0029 | |
| GSE77861 | RAPH1 | 65059 | 225188_at | 0.5407 | 0.0796 | |
| GSE97050 | RAPH1 | 65059 | A_33_P3217020 | 0.2063 | 0.4784 | |
| SRP007169 | RAPH1 | 65059 | RNAseq | 0.7041 | 0.1280 | |
| SRP008496 | RAPH1 | 65059 | RNAseq | 0.7066 | 0.0034 | |
| SRP064894 | RAPH1 | 65059 | RNAseq | -0.3850 | 0.2164 | |
| SRP133303 | RAPH1 | 65059 | RNAseq | -0.0159 | 0.9276 | |
| SRP159526 | RAPH1 | 65059 | RNAseq | -0.6896 | 0.0260 | |
| SRP193095 | RAPH1 | 65059 | RNAseq | -0.1522 | 0.3008 | |
| SRP219564 | RAPH1 | 65059 | RNAseq | -0.5809 | 0.1690 | |
| TCGA | RAPH1 | 65059 | RNAseq | -0.0723 | 0.2322 |
Upregulated datasets: 0; Downregulated datasets: 1.
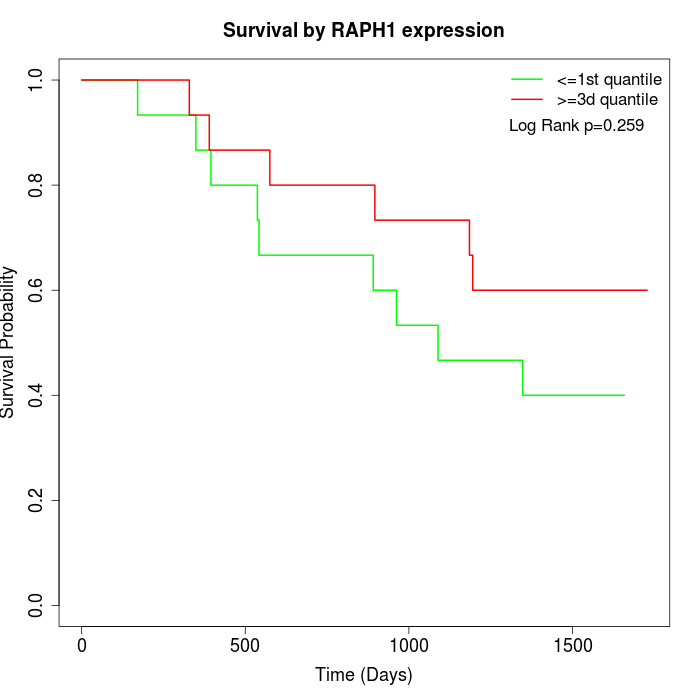
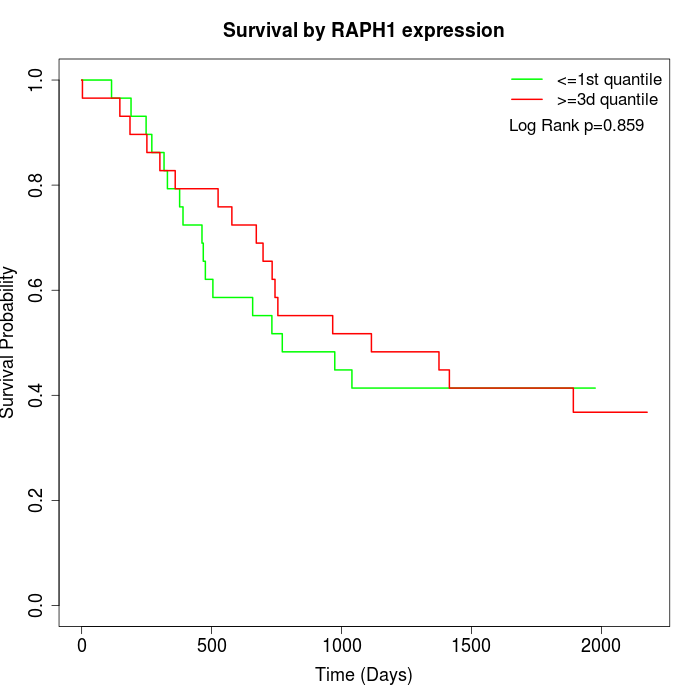
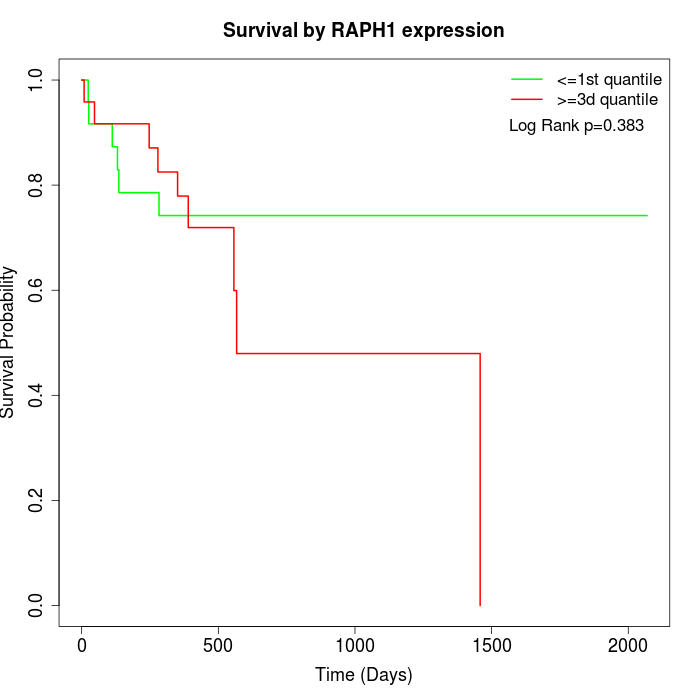
Survival by RAPH1 expression:
Note: Click image to view full size file.
Copy number change of RAPH1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAPH1 | 65059 | 7 | 4 | 19 | |
| GSE20123 | RAPH1 | 65059 | 7 | 4 | 19 | |
| GSE43470 | RAPH1 | 65059 | 2 | 4 | 37 | |
| GSE46452 | RAPH1 | 65059 | 1 | 4 | 54 | |
| GSE47630 | RAPH1 | 65059 | 4 | 5 | 31 | |
| GSE54993 | RAPH1 | 65059 | 0 | 3 | 67 | |
| GSE54994 | RAPH1 | 65059 | 10 | 8 | 35 | |
| GSE60625 | RAPH1 | 65059 | 0 | 3 | 8 | |
| GSE74703 | RAPH1 | 65059 | 2 | 3 | 31 | |
| GSE74704 | RAPH1 | 65059 | 3 | 2 | 15 | |
| TCGA | RAPH1 | 65059 | 20 | 13 | 63 |
Total number of gains: 56; Total number of losses: 53; Total Number of normals: 379.
Somatic mutations of RAPH1:
Generating mutation plots.
Highly correlated genes for RAPH1:
Showing top 20/317 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAPH1 | SLC25A15 | 0.780755 | 3 | 0 | 3 |
| RAPH1 | POPDC3 | 0.732595 | 3 | 0 | 3 |
| RAPH1 | PRRC2C | 0.725592 | 3 | 0 | 3 |
| RAPH1 | GORASP2 | 0.724644 | 3 | 0 | 3 |
| RAPH1 | DTYMK | 0.721907 | 3 | 0 | 3 |
| RAPH1 | CREB3L1 | 0.716447 | 3 | 0 | 3 |
| RAPH1 | AMIGO2 | 0.71433 | 3 | 0 | 3 |
| RAPH1 | DCAF13 | 0.707175 | 3 | 0 | 3 |
| RAPH1 | DAP3 | 0.692407 | 3 | 0 | 3 |
| RAPH1 | RBM15 | 0.688344 | 3 | 0 | 3 |
| RAPH1 | DDX10 | 0.680684 | 3 | 0 | 3 |
| RAPH1 | TMEM176A | 0.678303 | 3 | 0 | 3 |
| RAPH1 | ZNF281 | 0.677029 | 4 | 0 | 4 |
| RAPH1 | GTPBP8 | 0.67665 | 3 | 0 | 3 |
| RAPH1 | GLMN | 0.675559 | 3 | 0 | 3 |
| RAPH1 | KIF16B | 0.672022 | 3 | 0 | 3 |
| RAPH1 | SPIRE2 | 0.670598 | 3 | 0 | 3 |
| RAPH1 | INHBA | 0.670306 | 3 | 0 | 3 |
| RAPH1 | LIPG | 0.66904 | 3 | 0 | 3 |
| RAPH1 | CCT8 | 0.662403 | 3 | 0 | 3 |
For details and further investigation, click here