| Full name: inhibin subunit beta A | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7p14.1 | ||
| Entrez ID: 3624 | HGNC ID: HGNC:6066 | Ensembl Gene: ENSG00000122641 | OMIM ID: 147290 |
| Drug and gene relationship at DGIdb | |||
INHBA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04350 | TGF-beta signaling pathway | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells |
Expression of INHBA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | INHBA | 3624 | 227140_at | 3.1116 | 0.0599 | |
| GSE20347 | INHBA | 3624 | 210511_s_at | 2.8754 | 0.0000 | |
| GSE23400 | INHBA | 3624 | 210511_s_at | 2.0162 | 0.0000 | |
| GSE26886 | INHBA | 3624 | 210511_s_at | 4.4444 | 0.0000 | |
| GSE29001 | INHBA | 3624 | 210511_s_at | 3.4076 | 0.0000 | |
| GSE38129 | INHBA | 3624 | 210511_s_at | 2.6869 | 0.0000 | |
| GSE45670 | INHBA | 3624 | 227140_at | 2.0974 | 0.0674 | |
| GSE53622 | INHBA | 3624 | 35152 | 2.4832 | 0.0000 | |
| GSE53624 | INHBA | 3624 | 35152 | 2.9590 | 0.0000 | |
| GSE63941 | INHBA | 3624 | 210511_s_at | -4.3048 | 0.0046 | |
| GSE77861 | INHBA | 3624 | 210511_s_at | 1.3061 | 0.0505 | |
| GSE97050 | INHBA | 3624 | A_23_P122924 | 0.6193 | 0.1453 | |
| SRP007169 | INHBA | 3624 | RNAseq | 7.9898 | 0.0000 | |
| SRP008496 | INHBA | 3624 | RNAseq | 8.5692 | 0.0000 | |
| SRP064894 | INHBA | 3624 | RNAseq | 4.6921 | 0.0000 | |
| SRP133303 | INHBA | 3624 | RNAseq | 4.5513 | 0.0000 | |
| SRP159526 | INHBA | 3624 | RNAseq | 3.4111 | 0.0000 | |
| SRP193095 | INHBA | 3624 | RNAseq | 6.8563 | 0.0000 | |
| SRP219564 | INHBA | 3624 | RNAseq | 3.2517 | 0.0010 | |
| TCGA | INHBA | 3624 | RNAseq | 1.2556 | 0.0000 |
Upregulated datasets: 15; Downregulated datasets: 1.
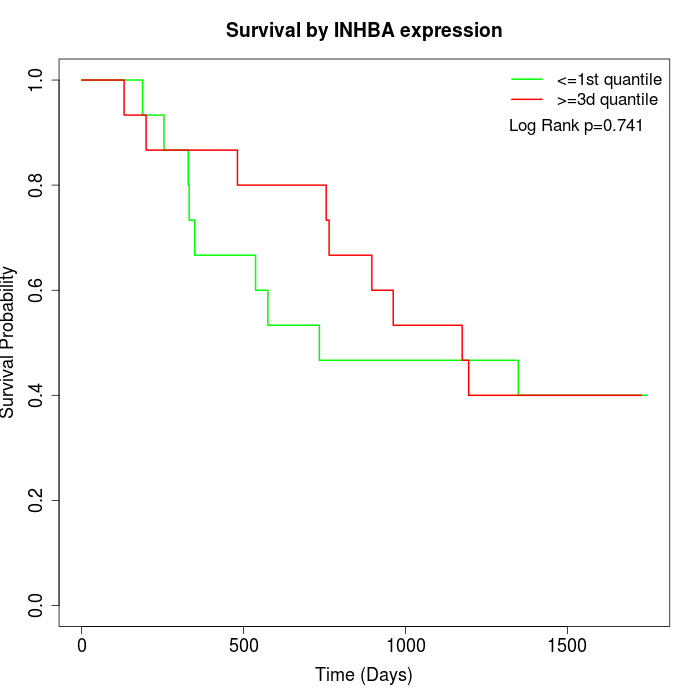
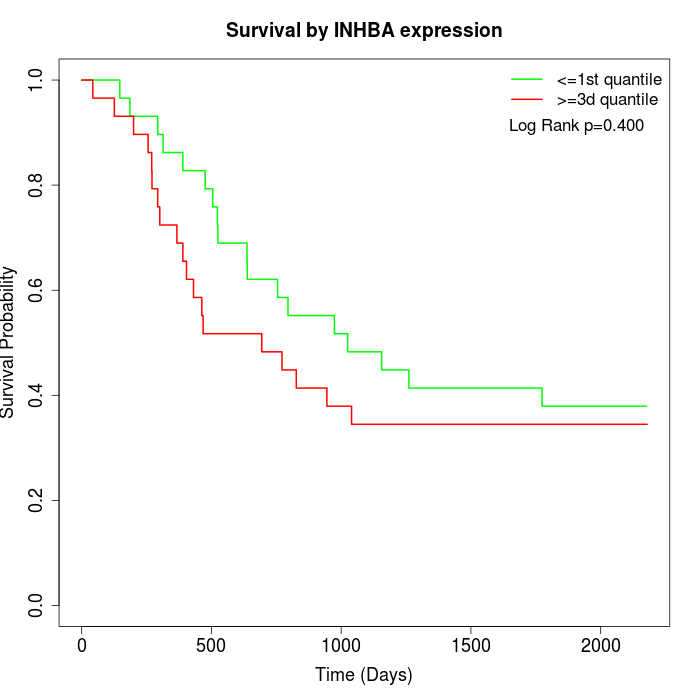
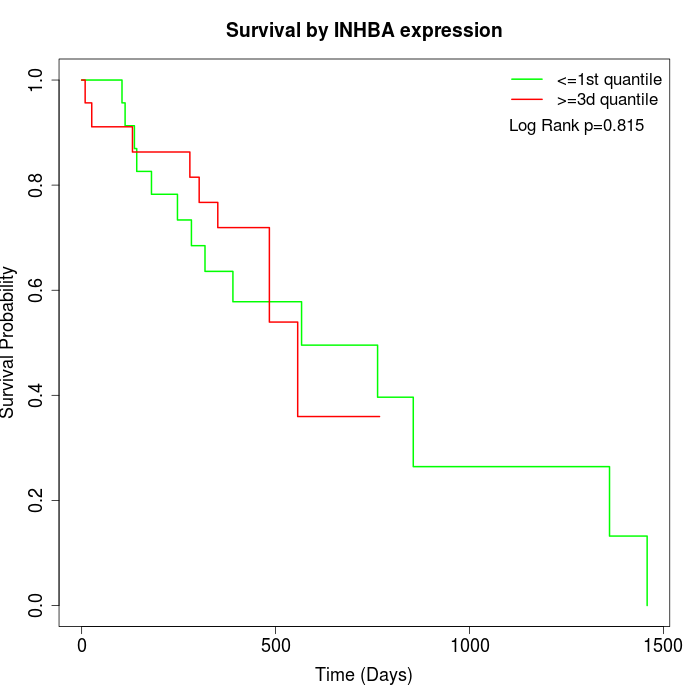
Survival by INHBA expression:
Note: Click image to view full size file.
Copy number change of INHBA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | INHBA | 3624 | 14 | 0 | 16 | |
| GSE20123 | INHBA | 3624 | 13 | 0 | 17 | |
| GSE43470 | INHBA | 3624 | 4 | 0 | 39 | |
| GSE46452 | INHBA | 3624 | 11 | 1 | 47 | |
| GSE47630 | INHBA | 3624 | 8 | 1 | 31 | |
| GSE54993 | INHBA | 3624 | 0 | 8 | 62 | |
| GSE54994 | INHBA | 3624 | 18 | 3 | 32 | |
| GSE60625 | INHBA | 3624 | 0 | 0 | 11 | |
| GSE74703 | INHBA | 3624 | 4 | 0 | 32 | |
| GSE74704 | INHBA | 3624 | 7 | 0 | 13 | |
| TCGA | INHBA | 3624 | 52 | 8 | 36 |
Total number of gains: 131; Total number of losses: 21; Total Number of normals: 336.
Somatic mutations of INHBA:
Generating mutation plots.
Highly correlated genes for INHBA:
Showing top 20/1266 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INHBA | PMEPA1 | 0.858908 | 10 | 0 | 10 |
| INHBA | LOXL2 | 0.849285 | 12 | 0 | 12 |
| INHBA | FAP | 0.83022 | 10 | 0 | 10 |
| INHBA | COL5A1 | 0.829458 | 12 | 0 | 12 |
| INHBA | MFAP2 | 0.826298 | 11 | 0 | 11 |
| INHBA | COL5A2 | 0.823186 | 12 | 0 | 12 |
| INHBA | TGFBI | 0.814919 | 12 | 0 | 12 |
| INHBA | WDR54 | 0.808172 | 6 | 0 | 6 |
| INHBA | COL1A1 | 0.801802 | 11 | 0 | 11 |
| INHBA | LGALS1 | 0.799374 | 12 | 0 | 11 |
| INHBA | BGN | 0.798996 | 12 | 0 | 12 |
| INHBA | VCAN | 0.798468 | 12 | 0 | 12 |
| INHBA | ENAH | 0.797028 | 11 | 0 | 10 |
| INHBA | LAMC2 | 0.79608 | 11 | 0 | 10 |
| INHBA | SERPINH1 | 0.79482 | 12 | 0 | 11 |
| INHBA | SNAI2 | 0.794126 | 8 | 0 | 8 |
| INHBA | POSTN | 0.787548 | 12 | 0 | 12 |
| INHBA | ITGAV | 0.786997 | 10 | 0 | 10 |
| INHBA | SPARC | 0.774413 | 12 | 0 | 12 |
| INHBA | SPP1 | 0.768101 | 11 | 0 | 10 |
For details and further investigation, click here