| Full name: ral guanine nucleotide dissociation stimulator like 2 | Alias Symbol: KE1.5|HKE1.5 | ||
| Type: protein-coding gene | Cytoband: 6p21.32 | ||
| Entrez ID: 5863 | HGNC ID: HGNC:9769 | Ensembl Gene: ENSG00000237441 | OMIM ID: 602306 |
| Drug and gene relationship at DGIdb | |||
RGL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway |
Expression of RGL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGL2 | 5863 | 209110_s_at | 0.0806 | 0.9121 | |
| GSE20347 | RGL2 | 5863 | 209110_s_at | -0.7042 | 0.0000 | |
| GSE23400 | RGL2 | 5863 | 209110_s_at | -0.4769 | 0.0000 | |
| GSE26886 | RGL2 | 5863 | 209110_s_at | 0.0003 | 0.9991 | |
| GSE29001 | RGL2 | 5863 | 209110_s_at | -0.8184 | 0.0009 | |
| GSE38129 | RGL2 | 5863 | 209110_s_at | -0.6380 | 0.0000 | |
| GSE45670 | RGL2 | 5863 | 209110_s_at | -0.2350 | 0.1859 | |
| GSE53622 | RGL2 | 5863 | 96519 | -0.6242 | 0.0000 | |
| GSE53624 | RGL2 | 5863 | 96519 | -0.6238 | 0.0000 | |
| GSE63941 | RGL2 | 5863 | 209110_s_at | 0.1945 | 0.6847 | |
| GSE77861 | RGL2 | 5863 | 209110_s_at | -0.5740 | 0.0305 | |
| GSE97050 | RGL2 | 5863 | A_23_P93383 | -0.2465 | 0.2330 | |
| SRP007169 | RGL2 | 5863 | RNAseq | -1.4149 | 0.0002 | |
| SRP008496 | RGL2 | 5863 | RNAseq | -1.2754 | 0.0000 | |
| SRP064894 | RGL2 | 5863 | RNAseq | -0.3918 | 0.0032 | |
| SRP133303 | RGL2 | 5863 | RNAseq | -0.8927 | 0.0000 | |
| SRP159526 | RGL2 | 5863 | RNAseq | -0.6873 | 0.0061 | |
| SRP193095 | RGL2 | 5863 | RNAseq | -0.4394 | 0.0007 | |
| SRP219564 | RGL2 | 5863 | RNAseq | -0.7006 | 0.0469 | |
| TCGA | RGL2 | 5863 | RNAseq | -0.1654 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 2.
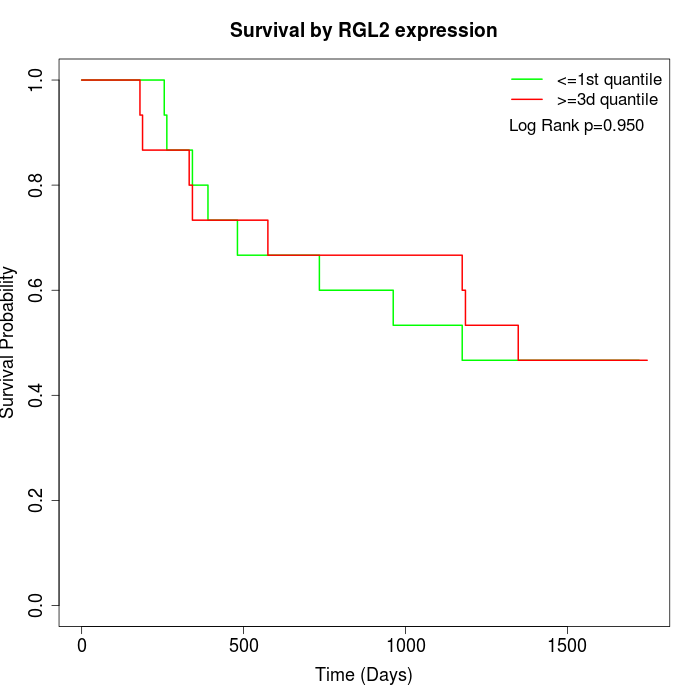
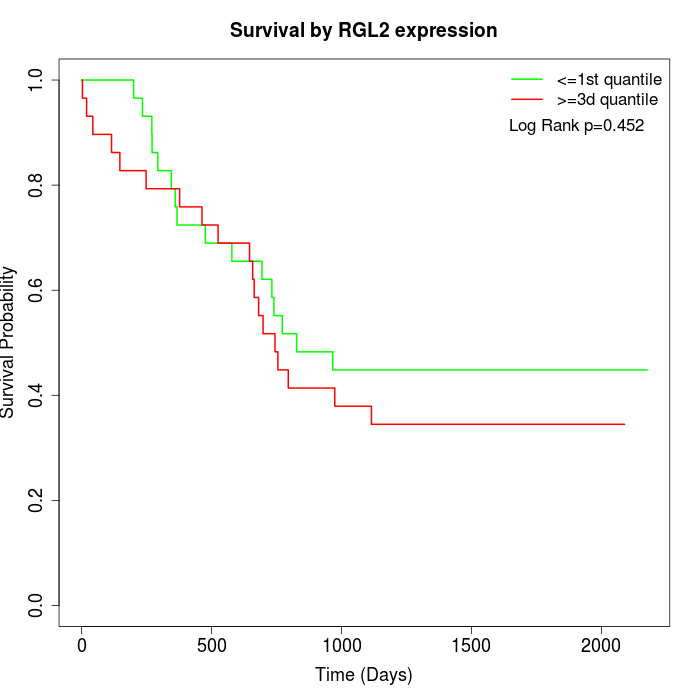
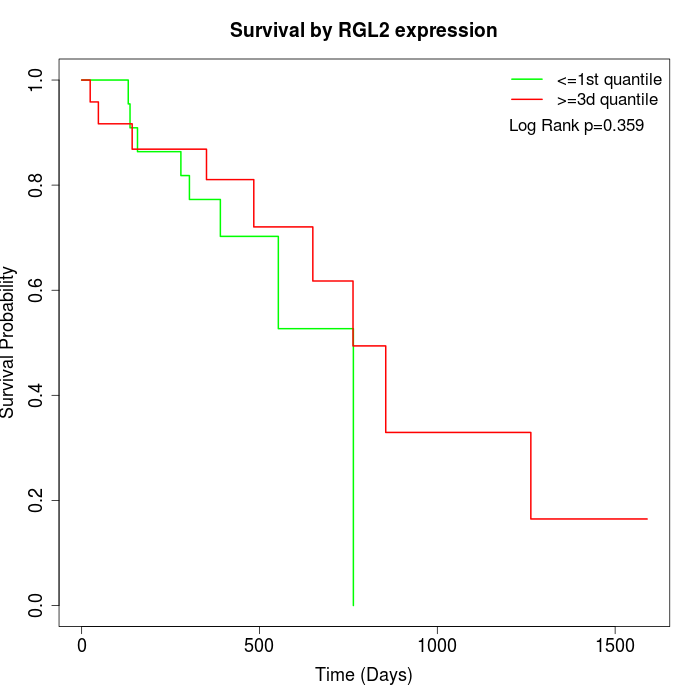
Survival by RGL2 expression:
Note: Click image to view full size file.
Copy number change of RGL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGL2 | 5863 | 5 | 0 | 25 | |
| GSE20123 | RGL2 | 5863 | 5 | 0 | 25 | |
| GSE43470 | RGL2 | 5863 | 4 | 1 | 38 | |
| GSE46452 | RGL2 | 5863 | 2 | 9 | 48 | |
| GSE47630 | RGL2 | 5863 | 8 | 4 | 28 | |
| GSE54993 | RGL2 | 5863 | 3 | 1 | 66 | |
| GSE54994 | RGL2 | 5863 | 11 | 4 | 38 | |
| GSE60625 | RGL2 | 5863 | 1 | 0 | 10 | |
| GSE74703 | RGL2 | 5863 | 4 | 0 | 32 | |
| GSE74704 | RGL2 | 5863 | 2 | 0 | 18 | |
| TCGA | RGL2 | 5863 | 15 | 16 | 65 |
Total number of gains: 60; Total number of losses: 35; Total Number of normals: 393.
Somatic mutations of RGL2:
Generating mutation plots.
Highly correlated genes for RGL2:
Showing top 20/807 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGL2 | TP53I3 | 0.718045 | 8 | 0 | 8 |
| RGL2 | TRIP10 | 0.704428 | 8 | 0 | 8 |
| RGL2 | CLIC3 | 0.700671 | 6 | 0 | 6 |
| RGL2 | ZNF600 | 0.69542 | 3 | 0 | 3 |
| RGL2 | SCNN1B | 0.692385 | 8 | 0 | 8 |
| RGL2 | NPAS2 | 0.684587 | 4 | 0 | 4 |
| RGL2 | ACOX3 | 0.683587 | 10 | 0 | 9 |
| RGL2 | CNPPD1 | 0.682498 | 8 | 0 | 7 |
| RGL2 | RAB5A | 0.681208 | 9 | 0 | 8 |
| RGL2 | KAT2B | 0.679294 | 8 | 0 | 7 |
| RGL2 | SORT1 | 0.67854 | 9 | 0 | 8 |
| RGL2 | FNDC4 | 0.67615 | 8 | 0 | 8 |
| RGL2 | VAV3 | 0.676006 | 6 | 0 | 5 |
| RGL2 | RANBP9 | 0.67562 | 8 | 0 | 8 |
| RGL2 | ALS2CL | 0.67511 | 9 | 0 | 8 |
| RGL2 | SYNGR1 | 0.672253 | 6 | 0 | 6 |
| RGL2 | FBXL16 | 0.67038 | 3 | 0 | 3 |
| RGL2 | BLNK | 0.670055 | 9 | 0 | 8 |
| RGL2 | ZNF185 | 0.669401 | 8 | 0 | 8 |
| RGL2 | SASH1 | 0.666096 | 9 | 0 | 8 |
For details and further investigation, click here