| Full name: regulator of G protein signaling 10 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q26.11 | ||
| Entrez ID: 6001 | HGNC ID: HGNC:9992 | Ensembl Gene: ENSG00000148908 | OMIM ID: 602856 |
| Drug and gene relationship at DGIdb | |||
Expression of RGS10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGS10 | 6001 | 204319_s_at | -0.3606 | 0.3967 | |
| GSE20347 | RGS10 | 6001 | 204319_s_at | -0.5800 | 0.0026 | |
| GSE23400 | RGS10 | 6001 | 204319_s_at | -0.3922 | 0.0001 | |
| GSE26886 | RGS10 | 6001 | 204319_s_at | -0.0742 | 0.8431 | |
| GSE29001 | RGS10 | 6001 | 204319_s_at | -0.6722 | 0.0259 | |
| GSE38129 | RGS10 | 6001 | 204319_s_at | -0.4216 | 0.0028 | |
| GSE45670 | RGS10 | 6001 | 204319_s_at | -0.3878 | 0.0275 | |
| GSE53622 | RGS10 | 6001 | 159928 | -0.3575 | 0.0001 | |
| GSE53624 | RGS10 | 6001 | 159928 | -0.4066 | 0.0000 | |
| GSE63941 | RGS10 | 6001 | 204319_s_at | -1.1468 | 0.0361 | |
| GSE77861 | RGS10 | 6001 | 204319_s_at | -0.8269 | 0.0483 | |
| GSE97050 | RGS10 | 6001 | A_23_P138717 | 0.0744 | 0.8362 | |
| SRP007169 | RGS10 | 6001 | RNAseq | -1.0004 | 0.0571 | |
| SRP008496 | RGS10 | 6001 | RNAseq | -1.1769 | 0.0003 | |
| SRP064894 | RGS10 | 6001 | RNAseq | -0.6713 | 0.0102 | |
| SRP133303 | RGS10 | 6001 | RNAseq | -0.3468 | 0.0640 | |
| SRP159526 | RGS10 | 6001 | RNAseq | -0.8104 | 0.0705 | |
| SRP193095 | RGS10 | 6001 | RNAseq | -0.6784 | 0.0001 | |
| SRP219564 | RGS10 | 6001 | RNAseq | -0.2194 | 0.4451 | |
| TCGA | RGS10 | 6001 | RNAseq | 0.2866 | 0.0006 |
Upregulated datasets: 0; Downregulated datasets: 2.
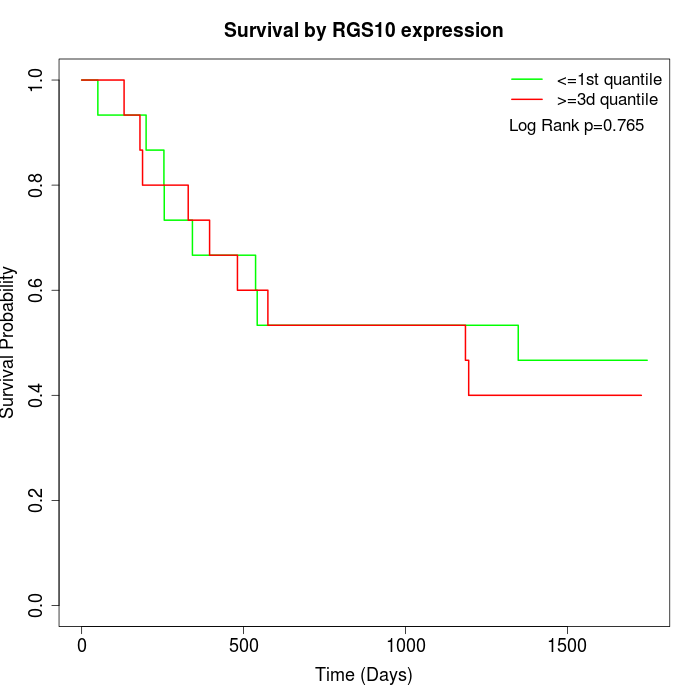
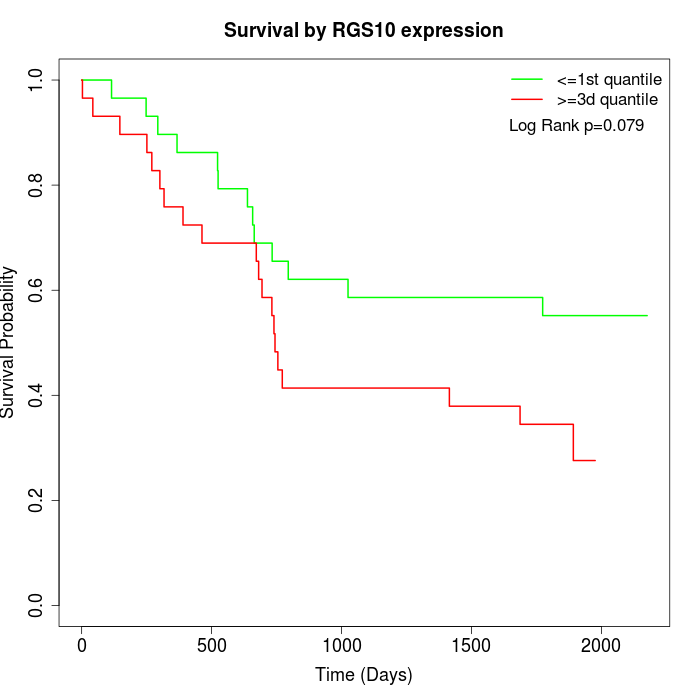
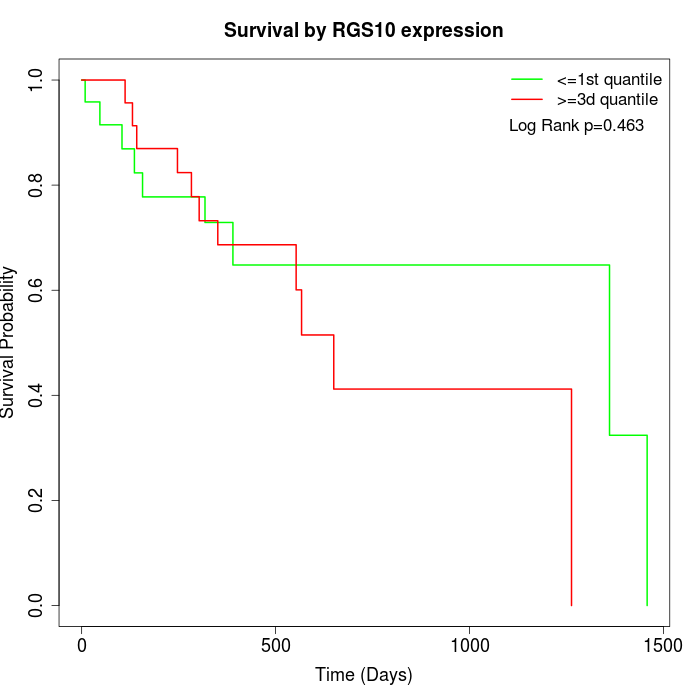
Survival by RGS10 expression:
Note: Click image to view full size file.
Copy number change of RGS10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGS10 | 6001 | 2 | 8 | 20 | |
| GSE20123 | RGS10 | 6001 | 2 | 8 | 20 | |
| GSE43470 | RGS10 | 6001 | 0 | 9 | 34 | |
| GSE46452 | RGS10 | 6001 | 0 | 11 | 48 | |
| GSE47630 | RGS10 | 6001 | 2 | 14 | 24 | |
| GSE54993 | RGS10 | 6001 | 8 | 0 | 62 | |
| GSE54994 | RGS10 | 6001 | 1 | 9 | 43 | |
| GSE60625 | RGS10 | 6001 | 0 | 0 | 11 | |
| GSE74703 | RGS10 | 6001 | 0 | 6 | 30 | |
| GSE74704 | RGS10 | 6001 | 1 | 3 | 16 | |
| TCGA | RGS10 | 6001 | 5 | 28 | 63 |
Total number of gains: 21; Total number of losses: 96; Total Number of normals: 371.
Somatic mutations of RGS10:
Generating mutation plots.
Highly correlated genes for RGS10:
Showing top 20/487 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGS10 | SS18 | 0.742174 | 3 | 0 | 3 |
| RGS10 | MBNL3 | 0.715214 | 4 | 0 | 3 |
| RGS10 | SVIP | 0.701035 | 3 | 0 | 3 |
| RGS10 | GTF2A2 | 0.690902 | 3 | 0 | 3 |
| RGS10 | RNF183 | 0.690028 | 3 | 0 | 3 |
| RGS10 | C3orf38 | 0.689349 | 4 | 0 | 4 |
| RGS10 | COMTD1 | 0.688716 | 4 | 0 | 3 |
| RGS10 | BRWD3 | 0.686362 | 3 | 0 | 3 |
| RGS10 | NIPSNAP3A | 0.680285 | 4 | 0 | 3 |
| RGS10 | ACAT1 | 0.679449 | 4 | 0 | 4 |
| RGS10 | ZNF407 | 0.679379 | 3 | 0 | 3 |
| RGS10 | DNAJC21 | 0.678467 | 3 | 0 | 3 |
| RGS10 | USP9Y | 0.676964 | 4 | 0 | 4 |
| RGS10 | ZNF626 | 0.67342 | 3 | 0 | 3 |
| RGS10 | BLOC1S2 | 0.66931 | 4 | 0 | 4 |
| RGS10 | OGFOD1 | 0.666183 | 3 | 0 | 3 |
| RGS10 | ACTR3C | 0.66411 | 4 | 0 | 4 |
| RGS10 | MBLAC2 | 0.658497 | 4 | 0 | 3 |
| RGS10 | PRKAG1 | 0.653451 | 3 | 0 | 3 |
| RGS10 | RBKS | 0.653217 | 4 | 0 | 3 |
For details and further investigation, click here