| Full name: RAS related | Alias Symbol: R-Ras | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 6237 | HGNC ID: HGNC:10447 | Ensembl Gene: ENSG00000126458 | OMIM ID: 165090 |
| Drug and gene relationship at DGIdb | |||
RRAS involved pathways:
Expression of RRAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRAS | 6237 | 212647_at | -0.3120 | 0.6958 | |
| GSE20347 | RRAS | 6237 | 212647_at | -0.1899 | 0.3429 | |
| GSE23400 | RRAS | 6237 | 212647_at | -0.1868 | 0.0791 | |
| GSE26886 | RRAS | 6237 | 212647_at | 0.4841 | 0.2103 | |
| GSE29001 | RRAS | 6237 | 212647_at | -0.0753 | 0.8174 | |
| GSE38129 | RRAS | 6237 | 212647_at | -0.6749 | 0.0564 | |
| GSE45670 | RRAS | 6237 | 212647_at | -0.4295 | 0.1128 | |
| GSE53622 | RRAS | 6237 | 72406 | -0.2780 | 0.0172 | |
| GSE53624 | RRAS | 6237 | 24222 | 0.1188 | 0.1987 | |
| GSE63941 | RRAS | 6237 | 212647_at | 0.1031 | 0.8794 | |
| GSE77861 | RRAS | 6237 | 212647_at | 0.3663 | 0.2965 | |
| GSE97050 | RRAS | 6237 | A_23_P39076 | -0.3078 | 0.5237 | |
| SRP007169 | RRAS | 6237 | RNAseq | -0.1948 | 0.6029 | |
| SRP008496 | RRAS | 6237 | RNAseq | -0.0254 | 0.9271 | |
| SRP064894 | RRAS | 6237 | RNAseq | -0.0870 | 0.7943 | |
| SRP133303 | RRAS | 6237 | RNAseq | 0.1843 | 0.5401 | |
| SRP159526 | RRAS | 6237 | RNAseq | -0.4404 | 0.0543 | |
| SRP193095 | RRAS | 6237 | RNAseq | 0.4879 | 0.0224 | |
| SRP219564 | RRAS | 6237 | RNAseq | -0.6145 | 0.4039 | |
| TCGA | RRAS | 6237 | RNAseq | -0.1413 | 0.0796 |
Upregulated datasets: 0; Downregulated datasets: 0.
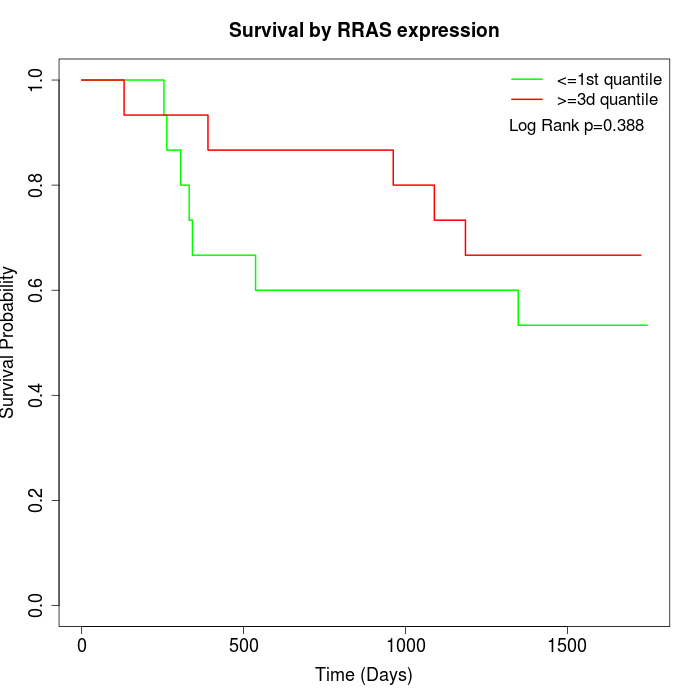
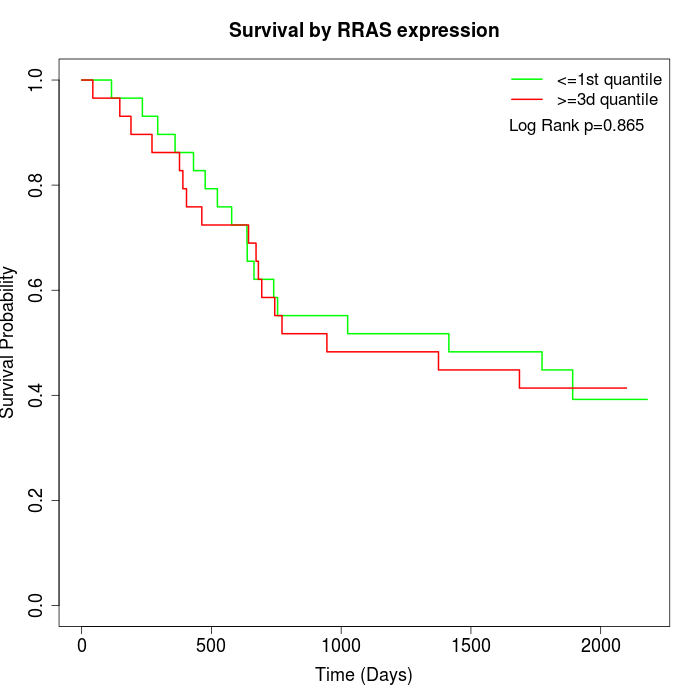
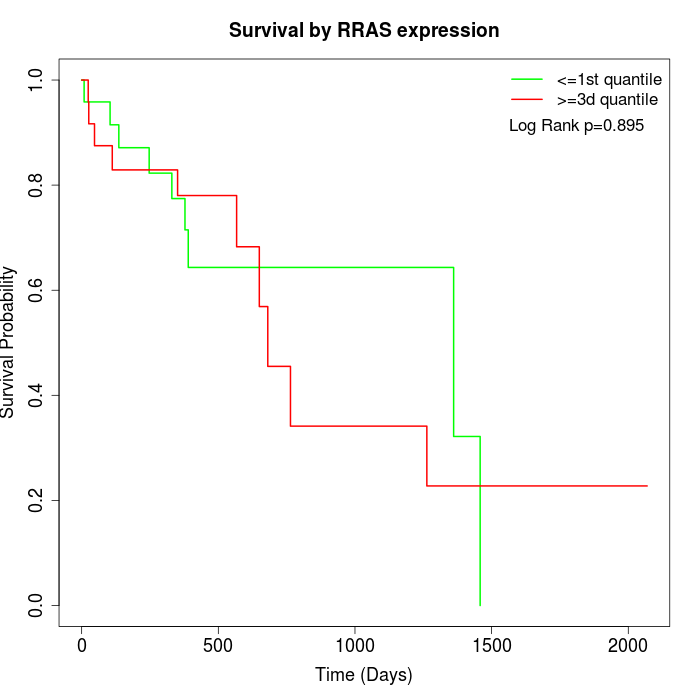
Survival by RRAS expression:
Note: Click image to view full size file.
Copy number change of RRAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RRAS | 6237 | 4 | 4 | 22 | |
| GSE20123 | RRAS | 6237 | 4 | 3 | 23 | |
| GSE43470 | RRAS | 6237 | 4 | 11 | 28 | |
| GSE46452 | RRAS | 6237 | 45 | 1 | 13 | |
| GSE47630 | RRAS | 6237 | 10 | 6 | 24 | |
| GSE54993 | RRAS | 6237 | 17 | 4 | 49 | |
| GSE54994 | RRAS | 6237 | 4 | 14 | 35 | |
| GSE60625 | RRAS | 6237 | 9 | 0 | 2 | |
| GSE74703 | RRAS | 6237 | 4 | 7 | 25 | |
| GSE74704 | RRAS | 6237 | 4 | 1 | 15 | |
| TCGA | RRAS | 6237 | 14 | 19 | 63 |
Total number of gains: 119; Total number of losses: 70; Total Number of normals: 299.
Somatic mutations of RRAS:
Generating mutation plots.
Highly correlated genes for RRAS:
Showing top 20/885 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRAS | DOCK11 | 0.790649 | 3 | 0 | 3 |
| RRAS | C20orf194 | 0.783335 | 3 | 0 | 3 |
| RRAS | C9orf47 | 0.772878 | 3 | 0 | 3 |
| RRAS | PLIN4 | 0.766906 | 3 | 0 | 3 |
| RRAS | ZNF540 | 0.761944 | 3 | 0 | 3 |
| RRAS | ITGB1BP2 | 0.760725 | 3 | 0 | 3 |
| RRAS | CD34 | 0.758619 | 5 | 0 | 5 |
| RRAS | SPOP | 0.758075 | 5 | 0 | 5 |
| RRAS | SOBP | 0.755412 | 6 | 0 | 6 |
| RRAS | ITGA7 | 0.752321 | 5 | 0 | 5 |
| RRAS | IQUB | 0.75021 | 3 | 0 | 3 |
| RRAS | MCAM | 0.747157 | 7 | 0 | 6 |
| RRAS | PCDH18 | 0.731204 | 3 | 0 | 3 |
| RRAS | ADARB1 | 0.728981 | 7 | 0 | 7 |
| RRAS | TUBG2 | 0.726329 | 4 | 0 | 4 |
| RRAS | SYNC | 0.724926 | 6 | 0 | 5 |
| RRAS | DUSP3 | 0.724099 | 5 | 0 | 5 |
| RRAS | FRY | 0.717864 | 4 | 0 | 3 |
| RRAS | CYS1 | 0.717398 | 6 | 0 | 6 |
| RRAS | ARHGEF25 | 0.714776 | 3 | 0 | 3 |
For details and further investigation, click here