| Full name: speckle type BTB/POZ protein | Alias Symbol: TEF2|BTBD32 | ||
| Type: protein-coding gene | Cytoband: 17q21.33 | ||
| Entrez ID: 8405 | HGNC ID: HGNC:11254 | Ensembl Gene: ENSG00000121067 | OMIM ID: 602650 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
SPOP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04340 | Hedgehog signaling pathway |
Expression of SPOP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPOP | 8405 | 204640_s_at | -0.6862 | 0.2846 | |
| GSE20347 | SPOP | 8405 | 204640_s_at | -0.0555 | 0.6795 | |
| GSE23400 | SPOP | 8405 | 204640_s_at | -0.2664 | 0.0235 | |
| GSE26886 | SPOP | 8405 | 204640_s_at | -0.8345 | 0.0011 | |
| GSE29001 | SPOP | 8405 | 208927_at | -0.1868 | 0.4379 | |
| GSE38129 | SPOP | 8405 | 204640_s_at | -0.3029 | 0.1089 | |
| GSE45670 | SPOP | 8405 | 208927_at | -0.3369 | 0.0054 | |
| GSE53622 | SPOP | 8405 | 56671 | -0.4200 | 0.0000 | |
| GSE53624 | SPOP | 8405 | 56671 | -0.1873 | 0.0013 | |
| GSE63941 | SPOP | 8405 | 204640_s_at | 0.9174 | 0.0140 | |
| GSE77861 | SPOP | 8405 | 204640_s_at | 0.1302 | 0.5797 | |
| GSE97050 | SPOP | 8405 | A_24_P270890 | -0.8310 | 0.0866 | |
| SRP007169 | SPOP | 8405 | RNAseq | -0.1709 | 0.5648 | |
| SRP008496 | SPOP | 8405 | RNAseq | -0.3088 | 0.0865 | |
| SRP064894 | SPOP | 8405 | RNAseq | -0.4729 | 0.0155 | |
| SRP133303 | SPOP | 8405 | RNAseq | -0.3456 | 0.0876 | |
| SRP159526 | SPOP | 8405 | RNAseq | -0.4751 | 0.0191 | |
| SRP193095 | SPOP | 8405 | RNAseq | -0.2267 | 0.0008 | |
| SRP219564 | SPOP | 8405 | RNAseq | -0.9860 | 0.1061 | |
| TCGA | SPOP | 8405 | RNAseq | -0.1943 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 0.
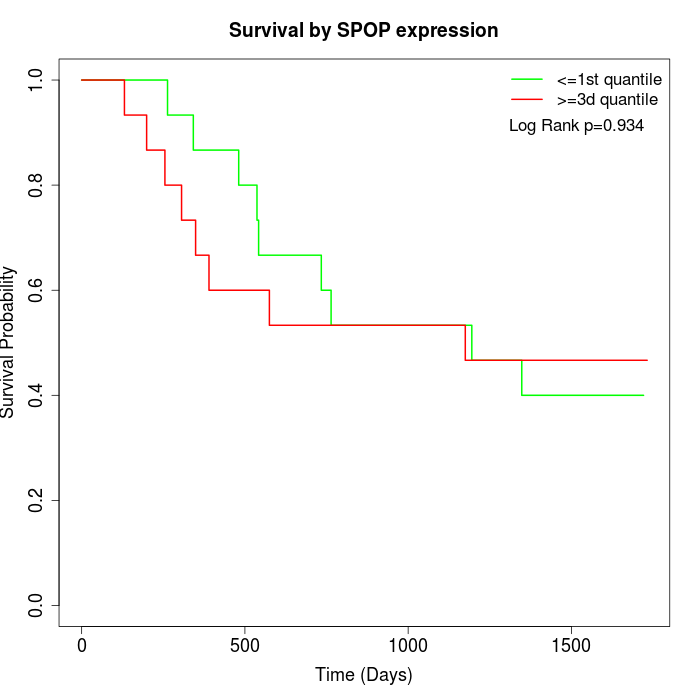
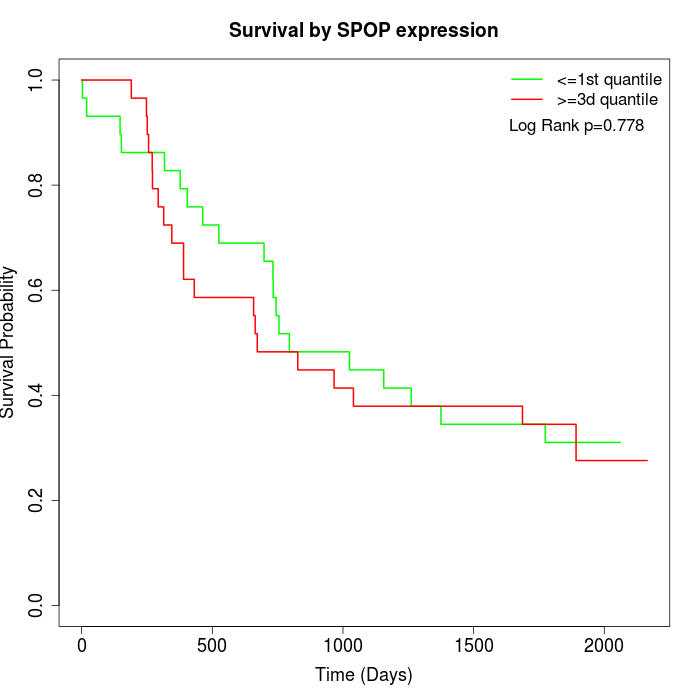
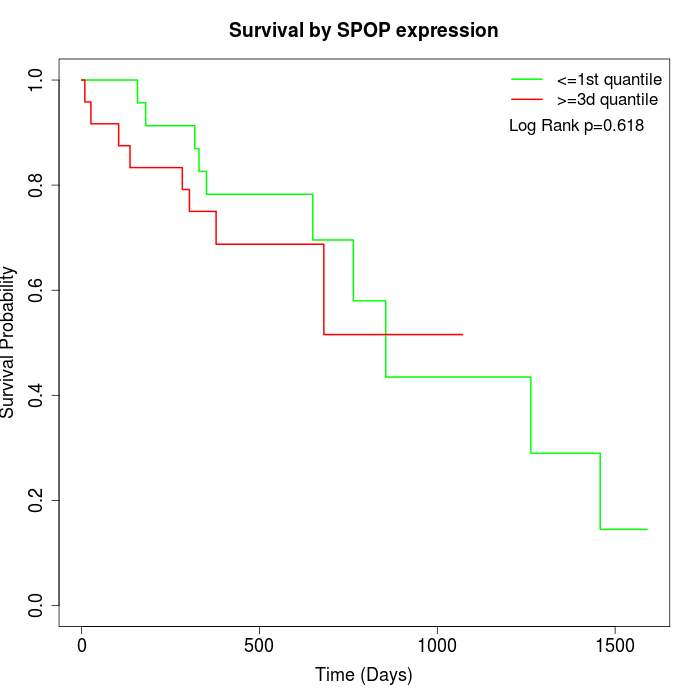
Survival by SPOP expression:
Note: Click image to view full size file.
Copy number change of SPOP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPOP | 8405 | 6 | 2 | 22 | |
| GSE20123 | SPOP | 8405 | 6 | 2 | 22 | |
| GSE43470 | SPOP | 8405 | 3 | 2 | 38 | |
| GSE46452 | SPOP | 8405 | 32 | 0 | 27 | |
| GSE47630 | SPOP | 8405 | 9 | 0 | 31 | |
| GSE54993 | SPOP | 8405 | 2 | 4 | 64 | |
| GSE54994 | SPOP | 8405 | 10 | 5 | 38 | |
| GSE60625 | SPOP | 8405 | 4 | 0 | 7 | |
| GSE74703 | SPOP | 8405 | 3 | 2 | 31 | |
| GSE74704 | SPOP | 8405 | 4 | 1 | 15 | |
| TCGA | SPOP | 8405 | 27 | 7 | 62 |
Total number of gains: 106; Total number of losses: 25; Total Number of normals: 357.
Somatic mutations of SPOP:
Generating mutation plots.
Highly correlated genes for SPOP:
Showing top 20/487 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPOP | PGR | 0.794786 | 4 | 0 | 4 |
| SPOP | CYS1 | 0.771178 | 4 | 0 | 4 |
| SPOP | ILK | 0.769076 | 6 | 0 | 5 |
| SPOP | PPARGC1A | 0.763494 | 6 | 0 | 6 |
| SPOP | A2M-AS1 | 0.759006 | 3 | 0 | 3 |
| SPOP | NCAM1 | 0.758966 | 6 | 0 | 6 |
| SPOP | RRAS | 0.758075 | 5 | 0 | 5 |
| SPOP | SLIT2 | 0.750019 | 6 | 0 | 6 |
| SPOP | CPXM2 | 0.749491 | 4 | 0 | 4 |
| SPOP | ITPR1 | 0.746482 | 7 | 0 | 7 |
| SPOP | KIAA1328 | 0.746089 | 3 | 0 | 3 |
| SPOP | MOAP1 | 0.746054 | 6 | 0 | 6 |
| SPOP | POLR3GL | 0.745281 | 4 | 0 | 4 |
| SPOP | PDK3 | 0.74154 | 4 | 0 | 4 |
| SPOP | PYGM | 0.740833 | 7 | 0 | 6 |
| SPOP | RNF115 | 0.7369 | 5 | 0 | 5 |
| SPOP | PTCHD1 | 0.735439 | 4 | 0 | 4 |
| SPOP | ANKS1B | 0.735028 | 3 | 0 | 3 |
| SPOP | CHN2 | 0.730299 | 3 | 0 | 3 |
| SPOP | SLC25A4 | 0.724062 | 7 | 0 | 6 |
For details and further investigation, click here