| Full name: ribonucleotide reductase catalytic subunit M1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11p15.4 | ||
| Entrez ID: 6240 | HGNC ID: HGNC:10451 | Ensembl Gene: ENSG00000167325 | OMIM ID: 180410 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RRM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRM1 | 6240 | 201477_s_at | 0.4975 | 0.3129 | |
| GSE20347 | RRM1 | 6240 | 201477_s_at | 0.6614 | 0.0004 | |
| GSE23400 | RRM1 | 6240 | 201477_s_at | 0.9865 | 0.0000 | |
| GSE26886 | RRM1 | 6240 | 201477_s_at | 0.8227 | 0.0001 | |
| GSE29001 | RRM1 | 6240 | 201477_s_at | 0.5621 | 0.0673 | |
| GSE38129 | RRM1 | 6240 | 201477_s_at | 0.8158 | 0.0000 | |
| GSE45670 | RRM1 | 6240 | 201477_s_at | 0.6701 | 0.0001 | |
| GSE53622 | RRM1 | 6240 | 153048 | 0.3892 | 0.0000 | |
| GSE53624 | RRM1 | 6240 | 153048 | 0.6229 | 0.0000 | |
| GSE63941 | RRM1 | 6240 | 201477_s_at | 0.8723 | 0.1105 | |
| GSE77861 | RRM1 | 6240 | 201477_s_at | 0.2538 | 0.5834 | |
| GSE97050 | RRM1 | 6240 | A_23_P87351 | 0.1924 | 0.5222 | |
| SRP007169 | RRM1 | 6240 | RNAseq | 0.7325 | 0.1159 | |
| SRP008496 | RRM1 | 6240 | RNAseq | 0.7856 | 0.0064 | |
| SRP064894 | RRM1 | 6240 | RNAseq | 0.4170 | 0.0439 | |
| SRP133303 | RRM1 | 6240 | RNAseq | 0.5121 | 0.0010 | |
| SRP159526 | RRM1 | 6240 | RNAseq | 0.3393 | 0.2519 | |
| SRP193095 | RRM1 | 6240 | RNAseq | 0.3113 | 0.0566 | |
| SRP219564 | RRM1 | 6240 | RNAseq | 0.0905 | 0.7674 | |
| TCGA | RRM1 | 6240 | RNAseq | 0.2572 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
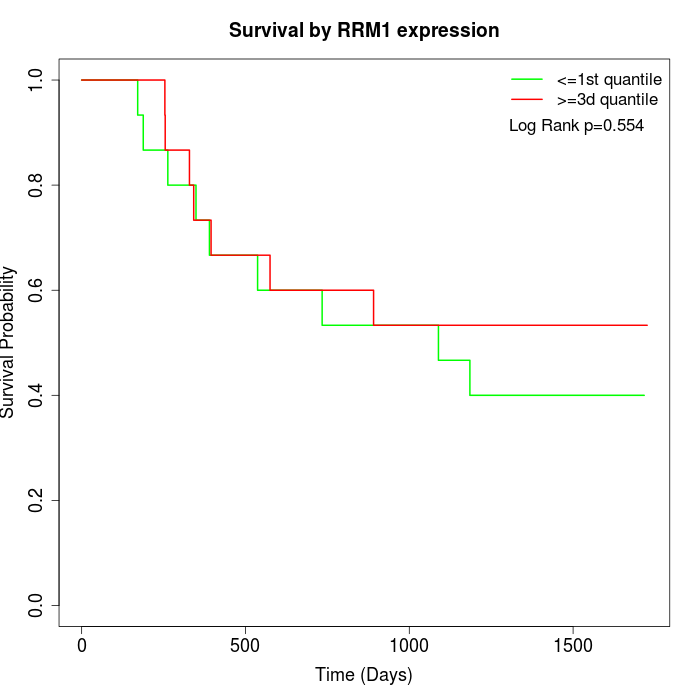
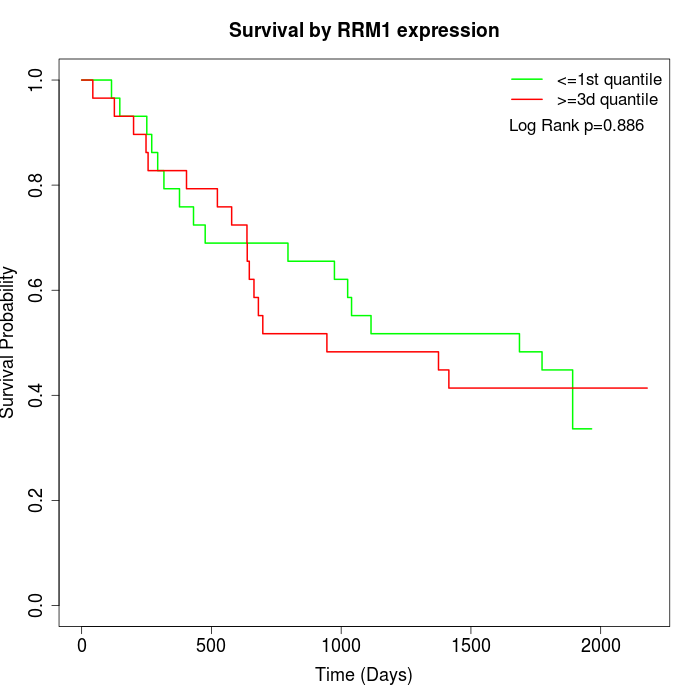
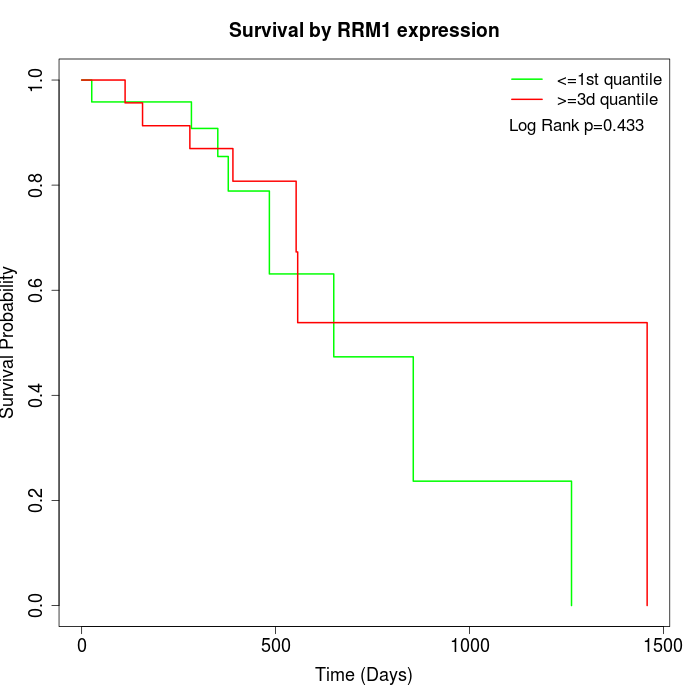
Survival by RRM1 expression:
Note: Click image to view full size file.
Copy number change of RRM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RRM1 | 6240 | 1 | 12 | 17 | |
| GSE20123 | RRM1 | 6240 | 1 | 11 | 18 | |
| GSE43470 | RRM1 | 6240 | 1 | 7 | 35 | |
| GSE46452 | RRM1 | 6240 | 7 | 8 | 44 | |
| GSE47630 | RRM1 | 6240 | 4 | 13 | 23 | |
| GSE54993 | RRM1 | 6240 | 3 | 1 | 66 | |
| GSE54994 | RRM1 | 6240 | 1 | 12 | 40 | |
| GSE60625 | RRM1 | 6240 | 0 | 0 | 11 | |
| GSE74703 | RRM1 | 6240 | 1 | 6 | 29 | |
| GSE74704 | RRM1 | 6240 | 0 | 8 | 12 | |
| TCGA | RRM1 | 6240 | 8 | 38 | 50 |
Total number of gains: 27; Total number of losses: 116; Total Number of normals: 345.
Somatic mutations of RRM1:
Generating mutation plots.
Highly correlated genes for RRM1:
Showing top 20/1434 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRM1 | NAA38 | 0.778558 | 3 | 0 | 3 |
| RRM1 | HRH1 | 0.762022 | 3 | 0 | 3 |
| RRM1 | ZNF607 | 0.743859 | 3 | 0 | 3 |
| RRM1 | PIK3R3 | 0.73202 | 3 | 0 | 3 |
| RRM1 | HDDC3 | 0.72326 | 3 | 0 | 3 |
| RRM1 | DKC1 | 0.712092 | 3 | 0 | 3 |
| RRM1 | NCAPG | 0.710298 | 10 | 0 | 9 |
| RRM1 | CENPL | 0.706982 | 7 | 0 | 7 |
| RRM1 | PSMG3 | 0.704944 | 6 | 0 | 6 |
| RRM1 | LRRC58 | 0.704057 | 4 | 0 | 4 |
| RRM1 | GLT8D1 | 0.703353 | 3 | 0 | 3 |
| RRM1 | CMC1 | 0.697977 | 3 | 0 | 3 |
| RRM1 | MELK | 0.6962 | 11 | 0 | 11 |
| RRM1 | CPSF3 | 0.693569 | 7 | 0 | 7 |
| RRM1 | MAD2L1 | 0.689369 | 9 | 0 | 9 |
| RRM1 | RMI1 | 0.688729 | 12 | 0 | 12 |
| RRM1 | GINS1 | 0.686887 | 12 | 0 | 11 |
| RRM1 | NDC80 | 0.68662 | 11 | 0 | 10 |
| RRM1 | ATR | 0.685568 | 10 | 0 | 10 |
| RRM1 | COLEC11 | 0.684157 | 3 | 0 | 3 |
For details and further investigation, click here