| Full name: mitotic arrest deficient 2 like 1 | Alias Symbol: MAD2|HSMAD2 | ||
| Type: protein-coding gene | Cytoband: 4q27 | ||
| Entrez ID: 4085 | HGNC ID: HGNC:6763 | Ensembl Gene: ENSG00000164109 | OMIM ID: 601467 |
| Drug and gene relationship at DGIdb | |||
MAD2L1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04914 | Progesterone-mediated oocyte maturation | |
| hsa05166 | HTLV-I infection |
Expression of MAD2L1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAD2L1 | 4085 | 203362_s_at | 1.5905 | 0.0248 | |
| GSE20347 | MAD2L1 | 4085 | 203362_s_at | 1.6525 | 0.0000 | |
| GSE23400 | MAD2L1 | 4085 | 203362_s_at | 1.3030 | 0.0000 | |
| GSE26886 | MAD2L1 | 4085 | 203362_s_at | 0.4085 | 0.2833 | |
| GSE29001 | MAD2L1 | 4085 | 203362_s_at | 1.5742 | 0.0164 | |
| GSE38129 | MAD2L1 | 4085 | 203362_s_at | 2.0785 | 0.0000 | |
| GSE45670 | MAD2L1 | 4085 | 203362_s_at | 1.1745 | 0.0000 | |
| GSE53622 | MAD2L1 | 4085 | 121299 | 1.4677 | 0.0000 | |
| GSE53624 | MAD2L1 | 4085 | 121299 | 1.6730 | 0.0000 | |
| GSE63941 | MAD2L1 | 4085 | 203362_s_at | 2.7200 | 0.0001 | |
| GSE77861 | MAD2L1 | 4085 | 203362_s_at | 1.0369 | 0.0405 | |
| GSE97050 | MAD2L1 | 4085 | A_23_P92441 | 0.6277 | 0.3488 | |
| SRP007169 | MAD2L1 | 4085 | RNAseq | 1.6000 | 0.0004 | |
| SRP008496 | MAD2L1 | 4085 | RNAseq | 1.3589 | 0.0000 | |
| SRP064894 | MAD2L1 | 4085 | RNAseq | 0.9125 | 0.0017 | |
| SRP133303 | MAD2L1 | 4085 | RNAseq | 1.6397 | 0.0000 | |
| SRP159526 | MAD2L1 | 4085 | RNAseq | 0.9039 | 0.0058 | |
| SRP193095 | MAD2L1 | 4085 | RNAseq | 0.6775 | 0.0000 | |
| SRP219564 | MAD2L1 | 4085 | RNAseq | 0.9186 | 0.1241 | |
| TCGA | MAD2L1 | 4085 | RNAseq | 1.0319 | 0.0000 |
Upregulated datasets: 14; Downregulated datasets: 0.
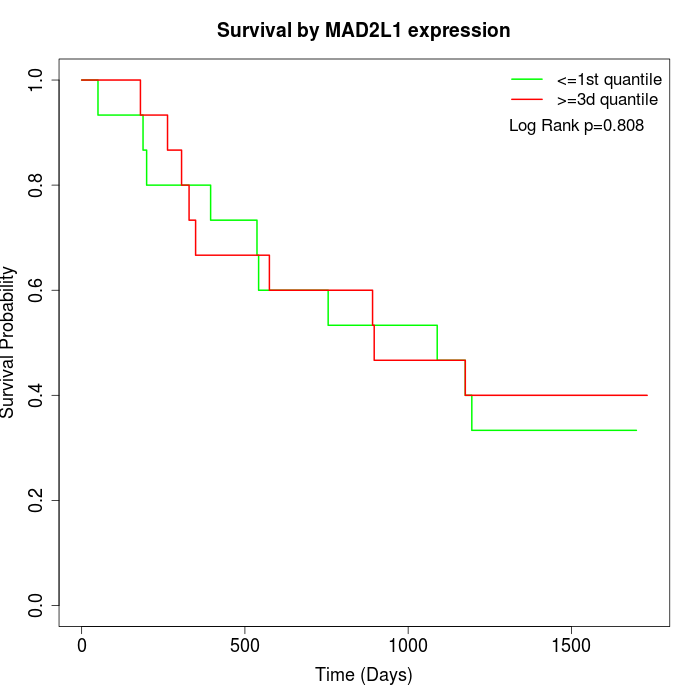
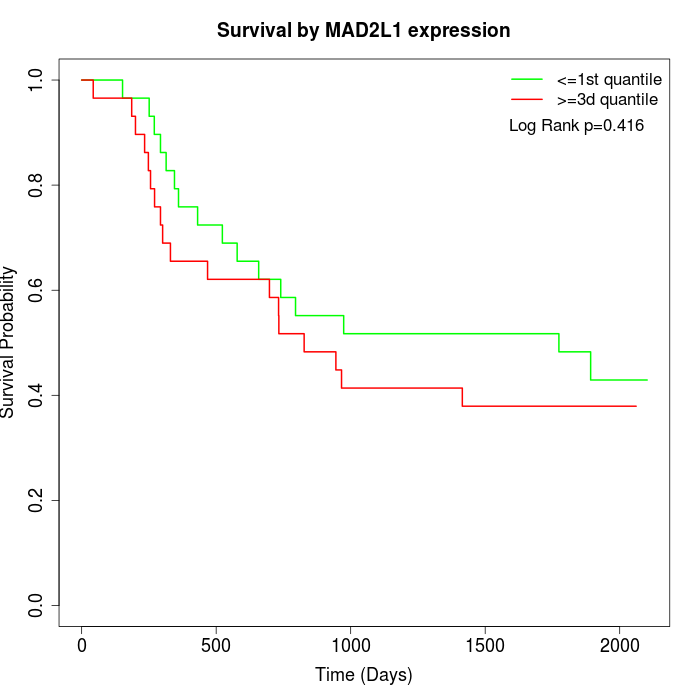
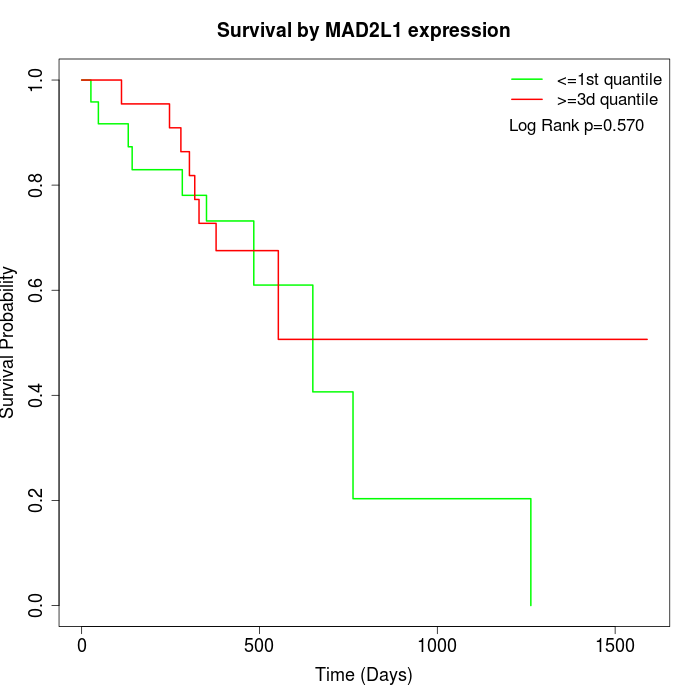
Survival by MAD2L1 expression:
Note: Click image to view full size file.
Copy number change of MAD2L1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAD2L1 | 4085 | 1 | 13 | 16 | |
| GSE20123 | MAD2L1 | 4085 | 1 | 13 | 16 | |
| GSE43470 | MAD2L1 | 4085 | 0 | 13 | 30 | |
| GSE46452 | MAD2L1 | 4085 | 1 | 36 | 22 | |
| GSE47630 | MAD2L1 | 4085 | 0 | 22 | 18 | |
| GSE54993 | MAD2L1 | 4085 | 9 | 0 | 61 | |
| GSE54994 | MAD2L1 | 4085 | 1 | 10 | 42 | |
| GSE60625 | MAD2L1 | 4085 | 0 | 1 | 10 | |
| GSE74703 | MAD2L1 | 4085 | 0 | 11 | 25 | |
| GSE74704 | MAD2L1 | 4085 | 0 | 7 | 13 | |
| TCGA | MAD2L1 | 4085 | 9 | 34 | 53 |
Total number of gains: 22; Total number of losses: 160; Total Number of normals: 306.
Somatic mutations of MAD2L1:
Generating mutation plots.
Highly correlated genes for MAD2L1:
Showing top 20/1612 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAD2L1 | CDK1 | 0.858199 | 12 | 0 | 12 |
| MAD2L1 | CCNA2 | 0.833401 | 13 | 0 | 12 |
| MAD2L1 | RAD51AP1 | 0.832361 | 13 | 0 | 13 |
| MAD2L1 | CCNB2 | 0.826999 | 11 | 0 | 11 |
| MAD2L1 | RACGAP1 | 0.817612 | 10 | 0 | 10 |
| MAD2L1 | ECT2 | 0.811368 | 13 | 0 | 12 |
| MAD2L1 | PBK | 0.802161 | 13 | 0 | 13 |
| MAD2L1 | PLK4 | 0.801666 | 12 | 0 | 12 |
| MAD2L1 | KIF2C | 0.800126 | 12 | 0 | 12 |
| MAD2L1 | CDCA5 | 0.798862 | 7 | 0 | 7 |
| MAD2L1 | KIF4A | 0.798516 | 12 | 0 | 12 |
| MAD2L1 | DLGAP5 | 0.795006 | 13 | 0 | 12 |
| MAD2L1 | MIS18A | 0.792699 | 8 | 0 | 8 |
| MAD2L1 | NEK2 | 0.791754 | 13 | 0 | 11 |
| MAD2L1 | SPC25 | 0.791003 | 12 | 0 | 11 |
| MAD2L1 | NUSAP1 | 0.790469 | 12 | 0 | 12 |
| MAD2L1 | MTFR2 | 0.788773 | 7 | 0 | 7 |
| MAD2L1 | MCM6 | 0.785713 | 12 | 0 | 12 |
| MAD2L1 | FOXM1 | 0.784745 | 12 | 0 | 12 |
| MAD2L1 | TTK | 0.783207 | 12 | 0 | 11 |
For details and further investigation, click here