| Full name: sterile alpha motif domain containing 15 | Alias Symbol: FLJ35963 | ||
| Type: protein-coding gene | Cytoband: 14q24.3 | ||
| Entrez ID: 161394 | HGNC ID: HGNC:18631 | Ensembl Gene: ENSG00000100583 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SAMD15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SAMD15 | 161394 | 1557680_at | -0.2847 | 0.2162 | |
| GSE26886 | SAMD15 | 161394 | 1557680_at | -0.4862 | 0.0581 | |
| GSE45670 | SAMD15 | 161394 | 1557680_at | 0.0987 | 0.5681 | |
| GSE53622 | SAMD15 | 161394 | 67402 | 1.3375 | 0.0000 | |
| GSE53624 | SAMD15 | 161394 | 67402 | 1.1952 | 0.0000 | |
| GSE63941 | SAMD15 | 161394 | 1557680_at | 0.3869 | 0.0695 | |
| GSE77861 | SAMD15 | 161394 | 1557680_at | -0.2384 | 0.1155 | |
| GSE97050 | SAMD15 | 161394 | A_23_P349025 | 0.2519 | 0.2500 | |
| SRP133303 | SAMD15 | 161394 | RNAseq | 1.6720 | 0.0000 | |
| SRP159526 | SAMD15 | 161394 | RNAseq | 1.8997 | 0.0002 |
Upregulated datasets: 4; Downregulated datasets: 0.
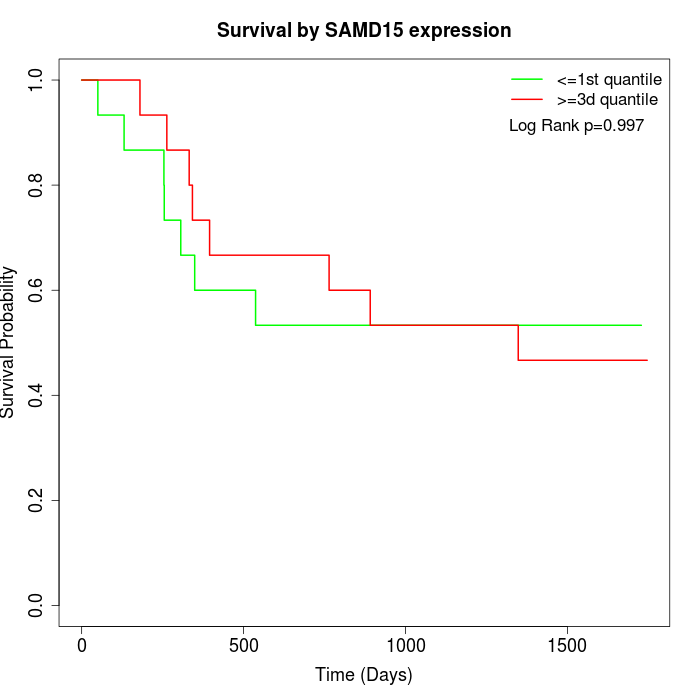
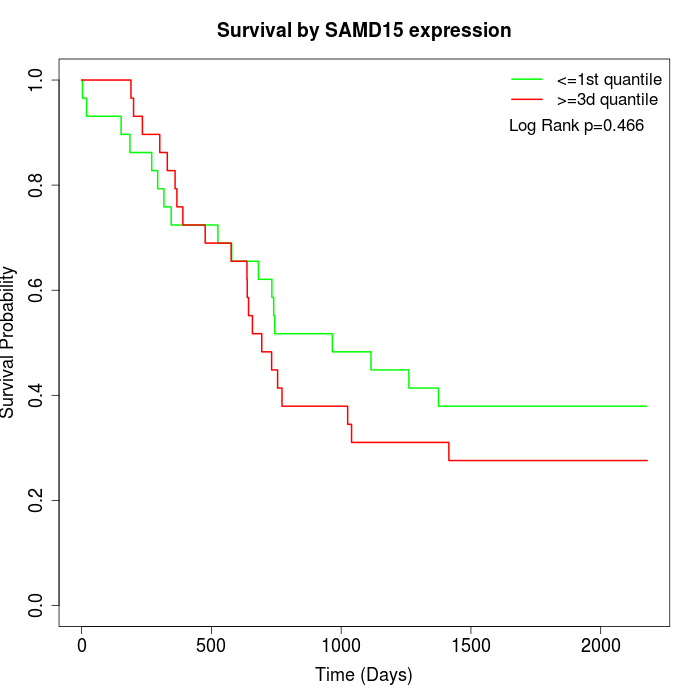
Survival by SAMD15 expression:
Note: Click image to view full size file.
Copy number change of SAMD15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SAMD15 | 161394 | 8 | 3 | 19 | |
| GSE20123 | SAMD15 | 161394 | 8 | 3 | 19 | |
| GSE43470 | SAMD15 | 161394 | 8 | 3 | 32 | |
| GSE46452 | SAMD15 | 161394 | 16 | 3 | 40 | |
| GSE47630 | SAMD15 | 161394 | 11 | 8 | 21 | |
| GSE54993 | SAMD15 | 161394 | 3 | 8 | 59 | |
| GSE54994 | SAMD15 | 161394 | 19 | 4 | 30 | |
| GSE60625 | SAMD15 | 161394 | 0 | 2 | 9 | |
| GSE74703 | SAMD15 | 161394 | 7 | 3 | 26 | |
| GSE74704 | SAMD15 | 161394 | 3 | 3 | 14 | |
| TCGA | SAMD15 | 161394 | 32 | 16 | 48 |
Total number of gains: 115; Total number of losses: 56; Total Number of normals: 317.
Somatic mutations of SAMD15:
Generating mutation plots.
Highly correlated genes for SAMD15:
Showing top 20/174 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SAMD15 | EMCN | 0.766501 | 3 | 0 | 3 |
| SAMD15 | MYCN | 0.726906 | 3 | 0 | 3 |
| SAMD15 | SIRPG | 0.719124 | 3 | 0 | 3 |
| SAMD15 | DMBT1 | 0.711994 | 3 | 0 | 3 |
| SAMD15 | CACNG6 | 0.704744 | 4 | 0 | 4 |
| SAMD15 | TOM1L1 | 0.699983 | 4 | 0 | 3 |
| SAMD15 | CYP2A7 | 0.699811 | 4 | 0 | 4 |
| SAMD15 | ZFPL1 | 0.697169 | 3 | 0 | 3 |
| SAMD15 | PZP | 0.695626 | 4 | 0 | 3 |
| SAMD15 | UBE2NL | 0.692853 | 3 | 0 | 3 |
| SAMD15 | MPV17L | 0.685286 | 4 | 0 | 3 |
| SAMD15 | APOB | 0.675497 | 4 | 0 | 4 |
| SAMD15 | TMC2 | 0.672496 | 5 | 0 | 4 |
| SAMD15 | IQCC | 0.669864 | 3 | 0 | 3 |
| SAMD15 | GNG10 | 0.668314 | 3 | 0 | 3 |
| SAMD15 | SLC4A8 | 0.662694 | 4 | 0 | 3 |
| SAMD15 | PLS1 | 0.662101 | 3 | 0 | 3 |
| SAMD15 | PRSS33 | 0.661618 | 3 | 0 | 3 |
| SAMD15 | MRGPRX4 | 0.659795 | 3 | 0 | 3 |
| SAMD15 | CSMD2-AS1 | 0.658572 | 3 | 0 | 3 |
For details and further investigation, click here