| Full name: semenogelin 2 | Alias Symbol: SGII | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 6407 | HGNC ID: HGNC:10743 | Ensembl Gene: ENSG00000124157 | OMIM ID: 182141 |
| Drug and gene relationship at DGIdb | |||
Expression of SEMG2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SEMG2 | 6407 | 216030_s_at | 0.0550 | 0.7922 | |
| GSE20347 | SEMG2 | 6407 | 216030_s_at | 0.1112 | 0.0735 | |
| GSE23400 | SEMG2 | 6407 | 216030_s_at | -0.0097 | 0.5919 | |
| GSE26886 | SEMG2 | 6407 | 216030_s_at | 0.0164 | 0.8749 | |
| GSE29001 | SEMG2 | 6407 | 216030_s_at | 0.0662 | 0.5169 | |
| GSE38129 | SEMG2 | 6407 | 216030_s_at | 0.1047 | 0.0579 | |
| GSE45670 | SEMG2 | 6407 | 216030_s_at | 0.0661 | 0.2921 | |
| GSE53622 | SEMG2 | 6407 | 13065 | 0.5547 | 0.0085 | |
| GSE53624 | SEMG2 | 6407 | 13065 | 0.5742 | 0.0002 | |
| GSE63941 | SEMG2 | 6407 | 216030_s_at | 0.0388 | 0.7603 | |
| GSE77861 | SEMG2 | 6407 | 216030_s_at | -0.0380 | 0.7055 | |
| GSE97050 | SEMG2 | 6407 | A_23_P5968 | 0.1292 | 0.5937 | |
| TCGA | SEMG2 | 6407 | RNAseq | 1.7392 | 0.3211 |
Upregulated datasets: 0; Downregulated datasets: 0.
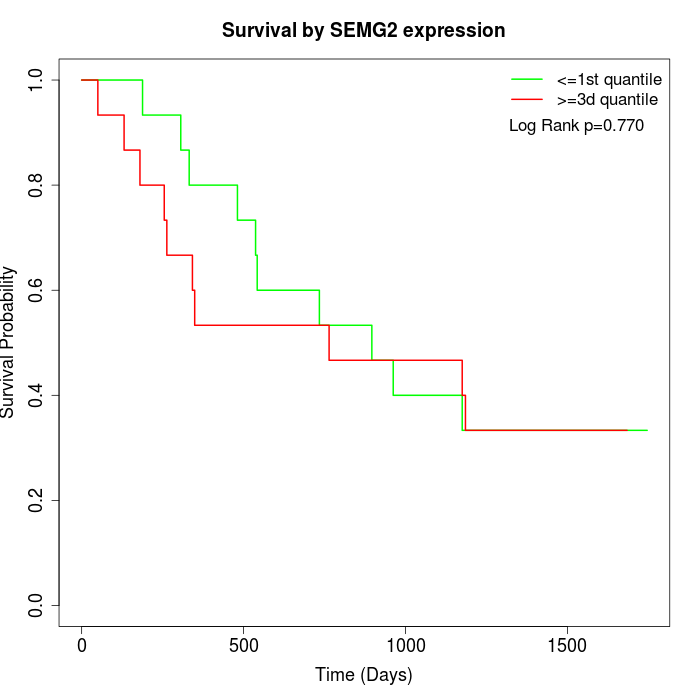
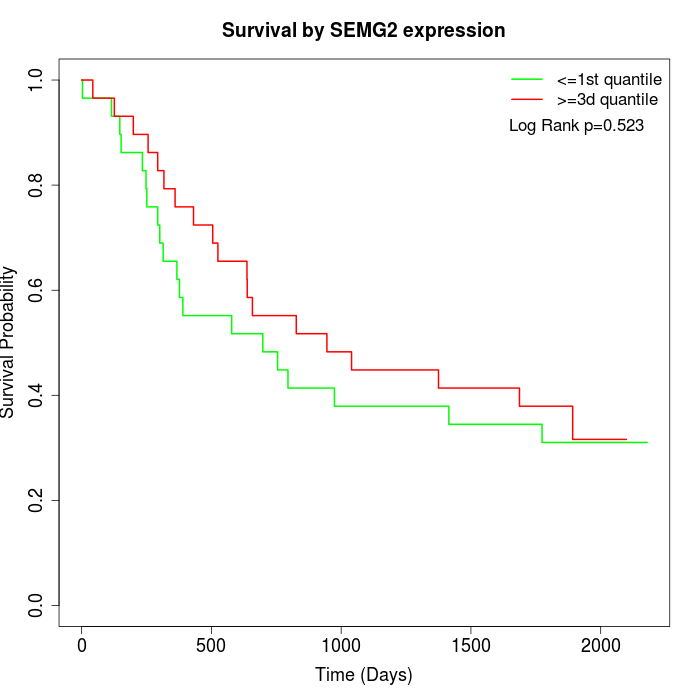
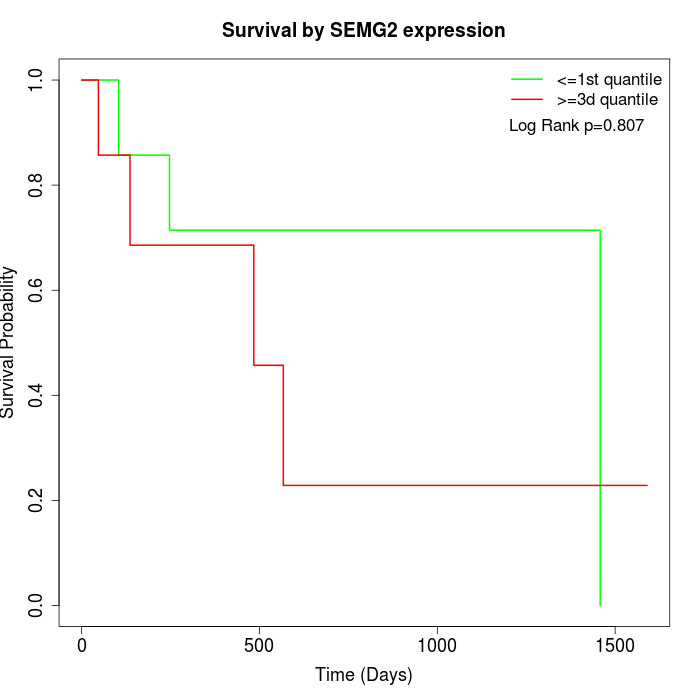
Survival by SEMG2 expression:
Note: Click image to view full size file.
Copy number change of SEMG2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SEMG2 | 6407 | 13 | 1 | 16 | |
| GSE20123 | SEMG2 | 6407 | 13 | 1 | 16 | |
| GSE43470 | SEMG2 | 6407 | 12 | 1 | 30 | |
| GSE46452 | SEMG2 | 6407 | 29 | 0 | 30 | |
| GSE47630 | SEMG2 | 6407 | 24 | 1 | 15 | |
| GSE54993 | SEMG2 | 6407 | 0 | 17 | 53 | |
| GSE54994 | SEMG2 | 6407 | 25 | 0 | 28 | |
| GSE60625 | SEMG2 | 6407 | 0 | 0 | 11 | |
| GSE74703 | SEMG2 | 6407 | 10 | 1 | 25 | |
| GSE74704 | SEMG2 | 6407 | 9 | 0 | 11 | |
| TCGA | SEMG2 | 6407 | 45 | 3 | 48 |
Total number of gains: 180; Total number of losses: 25; Total Number of normals: 283.
Somatic mutations of SEMG2:
Generating mutation plots.
Highly correlated genes for SEMG2:
Showing top 20/104 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SEMG2 | ZNF451 | 0.748202 | 3 | 0 | 3 |
| SEMG2 | ZNF780B | 0.703355 | 3 | 0 | 3 |
| SEMG2 | TEAD2 | 0.699213 | 3 | 0 | 3 |
| SEMG2 | IKZF3 | 0.697545 | 3 | 0 | 3 |
| SEMG2 | WDR86 | 0.685502 | 3 | 0 | 3 |
| SEMG2 | TRAF2 | 0.683873 | 4 | 0 | 3 |
| SEMG2 | FIP1L1 | 0.672827 | 3 | 0 | 3 |
| SEMG2 | GDI1 | 0.667954 | 3 | 0 | 3 |
| SEMG2 | RXRG | 0.665245 | 3 | 0 | 3 |
| SEMG2 | SH2B2 | 0.662439 | 5 | 0 | 4 |
| SEMG2 | GAL | 0.652832 | 4 | 0 | 3 |
| SEMG2 | KDM6B | 0.651581 | 4 | 0 | 3 |
| SEMG2 | DTX3 | 0.648886 | 3 | 0 | 3 |
| SEMG2 | NLGN4Y | 0.646061 | 3 | 0 | 3 |
| SEMG2 | HNRNPA1 | 0.642941 | 3 | 0 | 3 |
| SEMG2 | EVC2 | 0.63909 | 3 | 0 | 3 |
| SEMG2 | DNASE2 | 0.636943 | 3 | 0 | 3 |
| SEMG2 | ZNF683 | 0.636409 | 3 | 0 | 3 |
| SEMG2 | PRLR | 0.633348 | 3 | 0 | 3 |
| SEMG2 | ADAM22 | 0.624369 | 4 | 0 | 3 |
For details and further investigation, click here