| Full name: SET domain containing 2, histone lysine methyltransferase | Alias Symbol: HYPB|HIF-1|KIAA1732|FLJ23184|KMT3A | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 29072 | HGNC ID: HGNC:18420 | Ensembl Gene: ENSG00000181555 | OMIM ID: 612778 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SETD2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SETD2 | 29072 | 212493_s_at | -0.0969 | 0.7709 | |
| GSE20347 | SETD2 | 29072 | 212493_s_at | -0.3314 | 0.0009 | |
| GSE23400 | SETD2 | 29072 | 212493_s_at | -0.0348 | 0.3937 | |
| GSE26886 | SETD2 | 29072 | 212493_s_at | 0.5755 | 0.0010 | |
| GSE29001 | SETD2 | 29072 | 212493_s_at | -0.0470 | 0.8840 | |
| GSE38129 | SETD2 | 29072 | 212493_s_at | -0.3861 | 0.0002 | |
| GSE45670 | SETD2 | 29072 | 212493_s_at | -0.3491 | 0.0005 | |
| GSE53622 | SETD2 | 29072 | 16770 | -0.5177 | 0.0000 | |
| GSE53624 | SETD2 | 29072 | 16770 | -0.4477 | 0.0000 | |
| GSE63941 | SETD2 | 29072 | 212493_s_at | -0.0132 | 0.9627 | |
| GSE77861 | SETD2 | 29072 | 215038_s_at | -0.0612 | 0.7252 | |
| GSE97050 | SETD2 | 29072 | A_23_P44760 | -0.5541 | 0.1065 | |
| SRP007169 | SETD2 | 29072 | RNAseq | -0.2038 | 0.5732 | |
| SRP008496 | SETD2 | 29072 | RNAseq | -0.1571 | 0.4239 | |
| SRP064894 | SETD2 | 29072 | RNAseq | -0.5749 | 0.0085 | |
| SRP133303 | SETD2 | 29072 | RNAseq | -0.4312 | 0.0000 | |
| SRP159526 | SETD2 | 29072 | RNAseq | -0.5205 | 0.0008 | |
| SRP193095 | SETD2 | 29072 | RNAseq | -0.4297 | 0.0000 | |
| SRP219564 | SETD2 | 29072 | RNAseq | -0.4398 | 0.1125 | |
| TCGA | SETD2 | 29072 | RNAseq | -0.1448 | 0.0007 |
Upregulated datasets: 0; Downregulated datasets: 0.
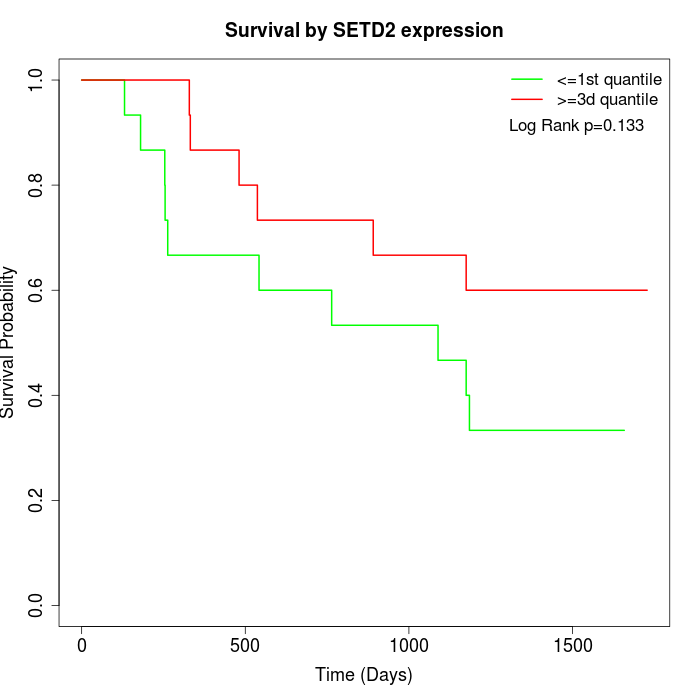
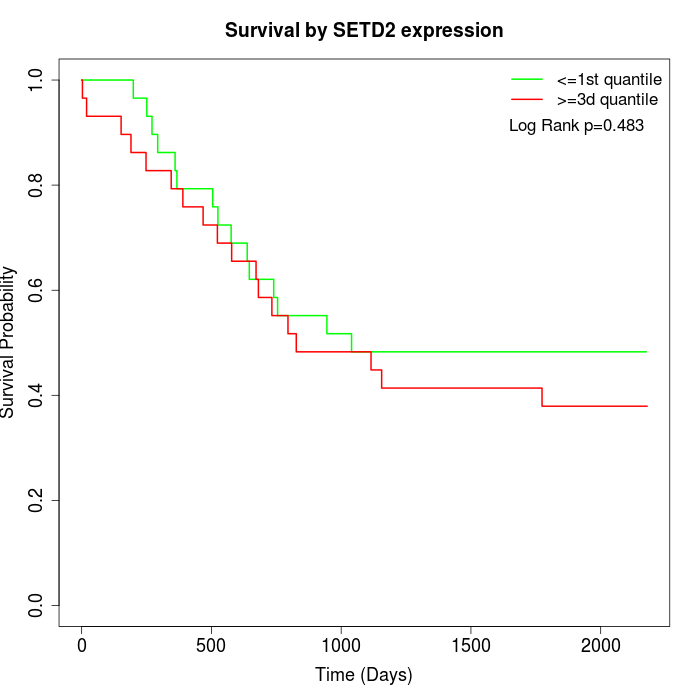
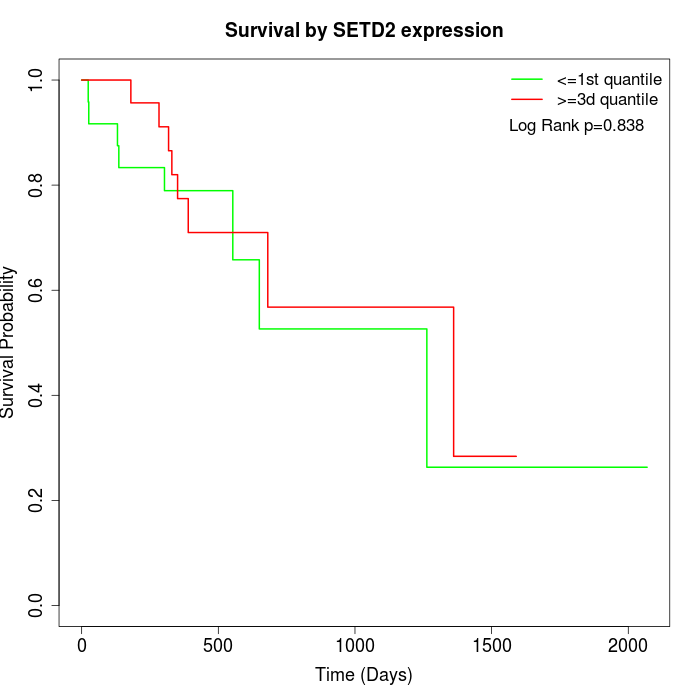
Survival by SETD2 expression:
Note: Click image to view full size file.
Copy number change of SETD2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SETD2 | 29072 | 0 | 18 | 12 | |
| GSE20123 | SETD2 | 29072 | 0 | 19 | 11 | |
| GSE43470 | SETD2 | 29072 | 0 | 19 | 24 | |
| GSE46452 | SETD2 | 29072 | 2 | 17 | 40 | |
| GSE47630 | SETD2 | 29072 | 2 | 23 | 15 | |
| GSE54993 | SETD2 | 29072 | 7 | 2 | 61 | |
| GSE54994 | SETD2 | 29072 | 0 | 36 | 17 | |
| GSE60625 | SETD2 | 29072 | 5 | 0 | 6 | |
| GSE74703 | SETD2 | 29072 | 0 | 15 | 21 | |
| GSE74704 | SETD2 | 29072 | 0 | 12 | 8 | |
| TCGA | SETD2 | 29072 | 1 | 75 | 20 |
Total number of gains: 17; Total number of losses: 236; Total Number of normals: 235.
Somatic mutations of SETD2:
Generating mutation plots.
Highly correlated genes for SETD2:
Showing top 20/529 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SETD2 | AGTPBP1 | 0.830806 | 3 | 0 | 3 |
| SETD2 | CDKL1 | 0.776098 | 3 | 0 | 3 |
| SETD2 | KLHDC1 | 0.772543 | 3 | 0 | 3 |
| SETD2 | NMNAT1 | 0.738521 | 3 | 0 | 3 |
| SETD2 | SUGT1 | 0.73059 | 3 | 0 | 3 |
| SETD2 | CLEC3B | 0.729678 | 3 | 0 | 3 |
| SETD2 | C11orf49 | 0.728147 | 3 | 0 | 3 |
| SETD2 | DYNC2H1 | 0.725637 | 3 | 0 | 3 |
| SETD2 | CYB5D1 | 0.724659 | 3 | 0 | 3 |
| SETD2 | KLHL15 | 0.723012 | 3 | 0 | 3 |
| SETD2 | PDS5B | 0.720607 | 3 | 0 | 3 |
| SETD2 | FYCO1 | 0.714319 | 6 | 0 | 5 |
| SETD2 | PBX2 | 0.711631 | 3 | 0 | 3 |
| SETD2 | FBXO8 | 0.70838 | 3 | 0 | 3 |
| SETD2 | ARL6IP5 | 0.700087 | 6 | 0 | 5 |
| SETD2 | POLK | 0.698401 | 3 | 0 | 3 |
| SETD2 | AP3S1 | 0.698065 | 3 | 0 | 3 |
| SETD2 | SACM1L | 0.697858 | 8 | 0 | 7 |
| SETD2 | KATNA1 | 0.697347 | 3 | 0 | 3 |
| SETD2 | AK4 | 0.697227 | 3 | 0 | 3 |
For details and further investigation, click here