| Full name: cathelicidin antimicrobial peptide | Alias Symbol: CAP18|FALL39|FALL-39|LL37 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 820 | HGNC ID: HGNC:1472 | Ensembl Gene: ENSG00000164047 | OMIM ID: 600474 |
| Drug and gene relationship at DGIdb | |||
CAMP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05152 | Tuberculosis |
Expression of CAMP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CAMP | 820 | 210244_at | 0.2424 | 0.4731 | |
| GSE20347 | CAMP | 820 | 210244_at | -0.0281 | 0.7196 | |
| GSE23400 | CAMP | 820 | 210244_at | -0.1189 | 0.0033 | |
| GSE26886 | CAMP | 820 | 210244_at | 0.1211 | 0.2509 | |
| GSE29001 | CAMP | 820 | 210244_at | -0.1581 | 0.3173 | |
| GSE38129 | CAMP | 820 | 210244_at | -0.0805 | 0.1884 | |
| GSE45670 | CAMP | 820 | 210244_at | 0.0518 | 0.6875 | |
| GSE53622 | CAMP | 820 | 5353 | 0.0618 | 0.8514 | |
| GSE53624 | CAMP | 820 | 5353 | -0.1273 | 0.5889 | |
| GSE63941 | CAMP | 820 | 210244_at | 0.2201 | 0.6062 | |
| GSE77861 | CAMP | 820 | 210244_at | -0.0125 | 0.9003 | |
| GSE97050 | CAMP | 820 | A_23_P253791 | 0.1125 | 0.5593 | |
| TCGA | CAMP | 820 | RNAseq | 0.5969 | 0.5401 |
Upregulated datasets: 0; Downregulated datasets: 0.
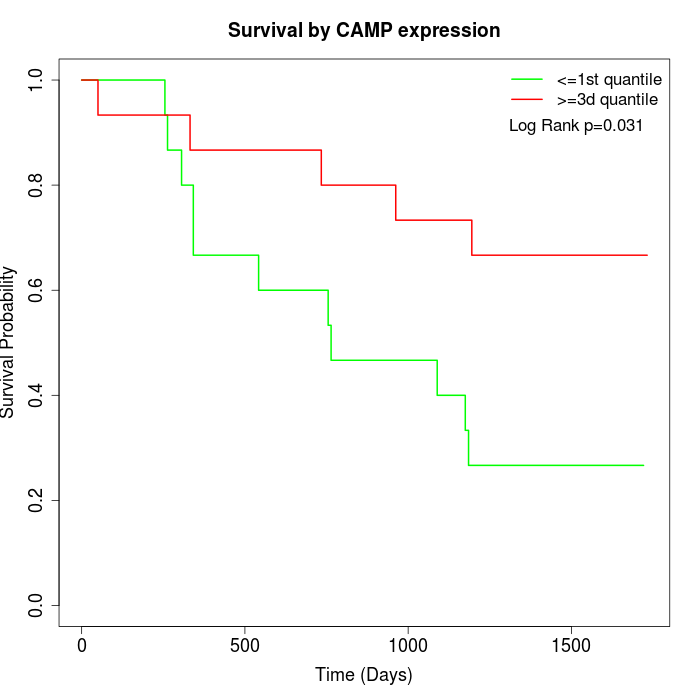
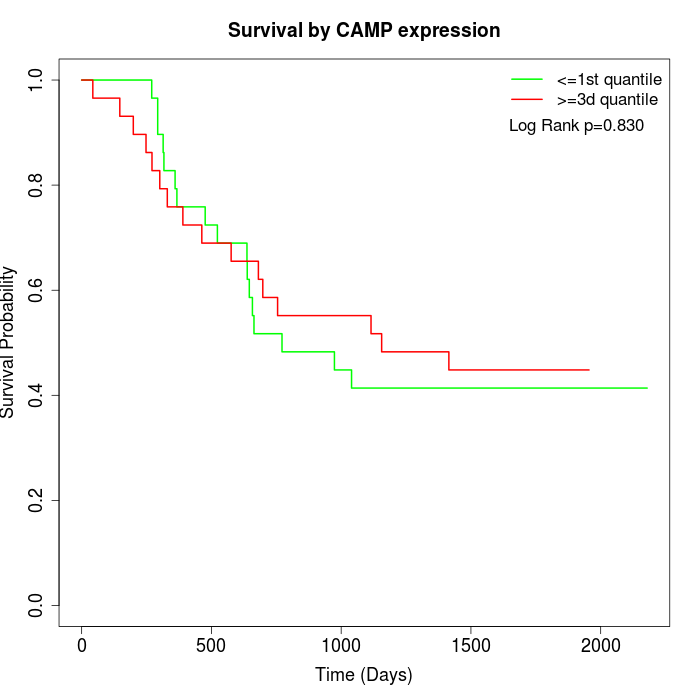
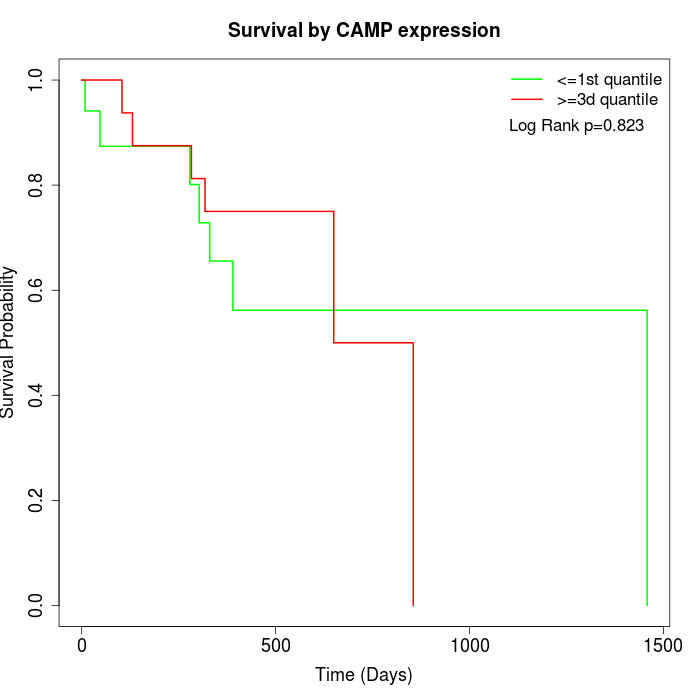
Survival by CAMP expression:
Note: Click image to view full size file.
Copy number change of CAMP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CAMP | 820 | 0 | 18 | 12 | |
| GSE20123 | CAMP | 820 | 0 | 19 | 11 | |
| GSE43470 | CAMP | 820 | 0 | 19 | 24 | |
| GSE46452 | CAMP | 820 | 2 | 17 | 40 | |
| GSE47630 | CAMP | 820 | 1 | 24 | 15 | |
| GSE54993 | CAMP | 820 | 6 | 2 | 62 | |
| GSE54994 | CAMP | 820 | 0 | 36 | 17 | |
| GSE60625 | CAMP | 820 | 5 | 0 | 6 | |
| GSE74703 | CAMP | 820 | 0 | 15 | 21 | |
| GSE74704 | CAMP | 820 | 0 | 12 | 8 | |
| TCGA | CAMP | 820 | 0 | 77 | 19 |
Total number of gains: 14; Total number of losses: 239; Total Number of normals: 235.
Somatic mutations of CAMP:
Generating mutation plots.
Highly correlated genes for CAMP:
Showing top 20/438 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CAMP | FBP2 | 0.697335 | 3 | 0 | 3 |
| CAMP | TTR | 0.697083 | 3 | 0 | 3 |
| CAMP | SNAI3 | 0.688452 | 3 | 0 | 3 |
| CAMP | SHBG | 0.672625 | 4 | 0 | 4 |
| CAMP | NLRC4 | 0.669121 | 4 | 0 | 3 |
| CAMP | FXR2 | 0.663576 | 3 | 0 | 3 |
| CAMP | RCVRN | 0.663543 | 6 | 0 | 5 |
| CAMP | SMPD3 | 0.658033 | 3 | 0 | 3 |
| CAMP | CXCR6 | 0.656343 | 3 | 0 | 3 |
| CAMP | MYH8 | 0.653953 | 3 | 0 | 3 |
| CAMP | KCNC4 | 0.647262 | 6 | 0 | 6 |
| CAMP | C8A | 0.639578 | 4 | 0 | 4 |
| CAMP | DHRSX | 0.639567 | 3 | 0 | 3 |
| CAMP | C11orf16 | 0.639485 | 5 | 0 | 4 |
| CAMP | CPT1C | 0.639456 | 3 | 0 | 3 |
| CAMP | EPHB1 | 0.63924 | 5 | 0 | 4 |
| CAMP | RFXAP | 0.639078 | 3 | 0 | 3 |
| CAMP | IL17A | 0.633467 | 5 | 0 | 5 |
| CAMP | HAVCR2 | 0.628992 | 3 | 0 | 3 |
| CAMP | TCTN2 | 0.628472 | 4 | 0 | 3 |
For details and further investigation, click here