| Full name: signal induced proliferation associated 1 like 1 | Alias Symbol: KIAA0440|E6TP1|SPAR1 | ||
| Type: protein-coding gene | Cytoband: 14q24.1 | ||
| Entrez ID: 26037 | HGNC ID: HGNC:20284 | Ensembl Gene: ENSG00000197555 | OMIM ID: 617504 |
| Drug and gene relationship at DGIdb | |||
SIPA1L1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway |
Expression of SIPA1L1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SIPA1L1 | 26037 | 202255_s_at | -0.0421 | 0.8879 | |
| GSE20347 | SIPA1L1 | 26037 | 202255_s_at | -0.0435 | 0.7683 | |
| GSE23400 | SIPA1L1 | 26037 | 202255_s_at | -0.0503 | 0.3612 | |
| GSE26886 | SIPA1L1 | 26037 | 202255_s_at | 0.7878 | 0.0003 | |
| GSE29001 | SIPA1L1 | 26037 | 202255_s_at | -0.0671 | 0.7675 | |
| GSE38129 | SIPA1L1 | 26037 | 202255_s_at | -0.0096 | 0.9358 | |
| GSE45670 | SIPA1L1 | 26037 | 202255_s_at | 0.2562 | 0.0578 | |
| GSE53622 | SIPA1L1 | 26037 | 147626 | 0.1672 | 0.0219 | |
| GSE53624 | SIPA1L1 | 26037 | 147626 | 0.1802 | 0.0007 | |
| GSE63941 | SIPA1L1 | 26037 | 202255_s_at | -0.6097 | 0.1807 | |
| GSE77861 | SIPA1L1 | 26037 | 202255_s_at | 0.1594 | 0.5897 | |
| GSE97050 | SIPA1L1 | 26037 | A_23_P76969 | 0.1136 | 0.6732 | |
| SRP007169 | SIPA1L1 | 26037 | RNAseq | 1.0250 | 0.0035 | |
| SRP008496 | SIPA1L1 | 26037 | RNAseq | 1.1874 | 0.0001 | |
| SRP064894 | SIPA1L1 | 26037 | RNAseq | 0.0655 | 0.6767 | |
| SRP133303 | SIPA1L1 | 26037 | RNAseq | 0.3281 | 0.0419 | |
| SRP159526 | SIPA1L1 | 26037 | RNAseq | -0.1432 | 0.6997 | |
| SRP193095 | SIPA1L1 | 26037 | RNAseq | 0.1627 | 0.2163 | |
| SRP219564 | SIPA1L1 | 26037 | RNAseq | -0.1736 | 0.6536 | |
| TCGA | SIPA1L1 | 26037 | RNAseq | -0.1450 | 0.0017 |
Upregulated datasets: 2; Downregulated datasets: 0.
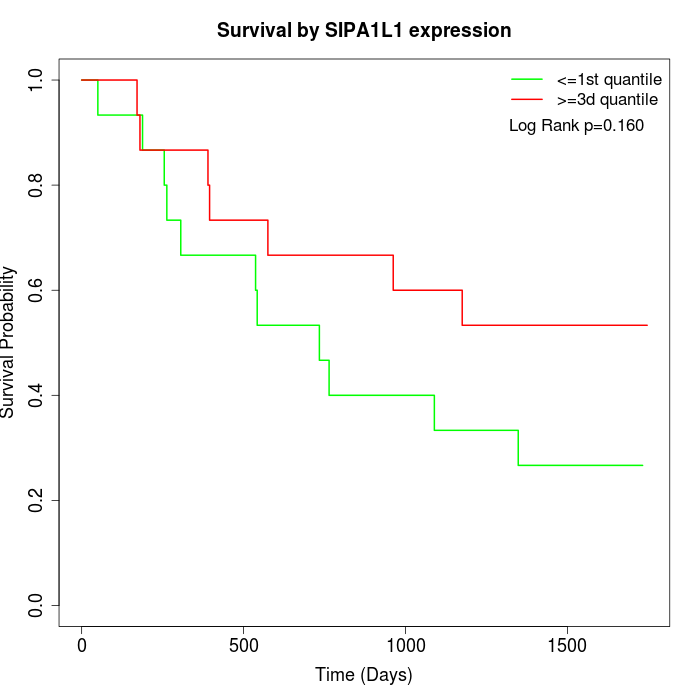
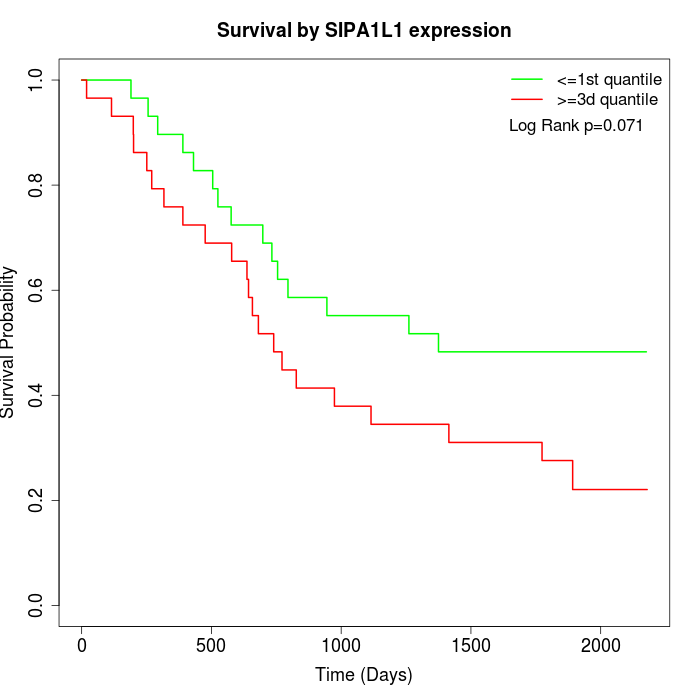
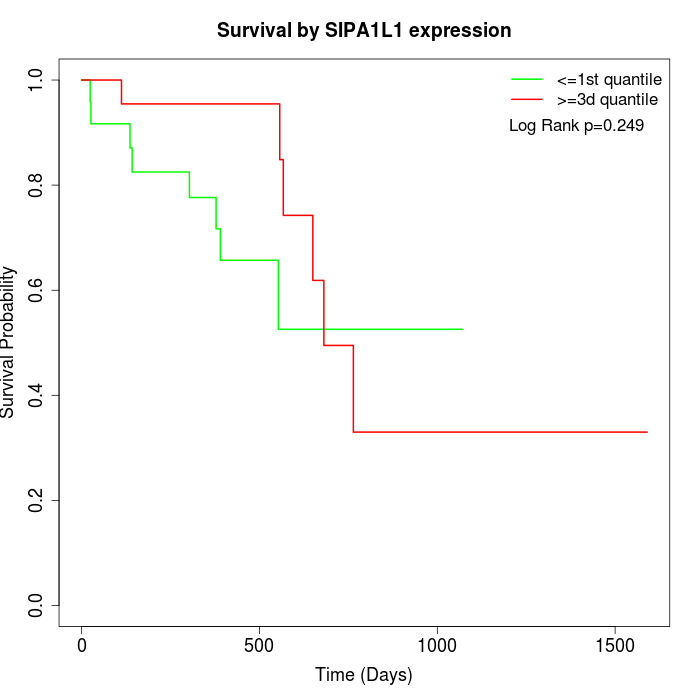
Survival by SIPA1L1 expression:
Note: Click image to view full size file.
Copy number change of SIPA1L1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SIPA1L1 | 26037 | 8 | 3 | 19 | |
| GSE20123 | SIPA1L1 | 26037 | 8 | 3 | 19 | |
| GSE43470 | SIPA1L1 | 26037 | 8 | 3 | 32 | |
| GSE46452 | SIPA1L1 | 26037 | 16 | 3 | 40 | |
| GSE47630 | SIPA1L1 | 26037 | 11 | 9 | 20 | |
| GSE54993 | SIPA1L1 | 26037 | 3 | 9 | 58 | |
| GSE54994 | SIPA1L1 | 26037 | 18 | 5 | 30 | |
| GSE60625 | SIPA1L1 | 26037 | 0 | 2 | 9 | |
| GSE74703 | SIPA1L1 | 26037 | 7 | 3 | 26 | |
| GSE74704 | SIPA1L1 | 26037 | 3 | 3 | 14 | |
| TCGA | SIPA1L1 | 26037 | 32 | 17 | 47 |
Total number of gains: 114; Total number of losses: 60; Total Number of normals: 314.
Somatic mutations of SIPA1L1:
Generating mutation plots.
Highly correlated genes for SIPA1L1:
Showing top 20/54 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SIPA1L1 | CCDC130 | 0.725625 | 3 | 0 | 3 |
| SIPA1L1 | MLF1 | 0.680609 | 3 | 0 | 3 |
| SIPA1L1 | RPA4 | 0.677212 | 4 | 0 | 4 |
| SIPA1L1 | F2RL2 | 0.661416 | 3 | 0 | 3 |
| SIPA1L1 | SUPT6H | 0.656835 | 3 | 0 | 3 |
| SIPA1L1 | GTF2IRD2 | 0.653403 | 4 | 0 | 4 |
| SIPA1L1 | OPN3 | 0.63073 | 3 | 0 | 3 |
| SIPA1L1 | NOB1 | 0.622961 | 3 | 0 | 3 |
| SIPA1L1 | AMPD2 | 0.621298 | 4 | 0 | 3 |
| SIPA1L1 | CLPB | 0.621278 | 3 | 0 | 3 |
| SIPA1L1 | YDJC | 0.620893 | 4 | 0 | 3 |
| SIPA1L1 | NPW | 0.615102 | 3 | 0 | 3 |
| SIPA1L1 | ZSCAN5A | 0.60909 | 3 | 0 | 3 |
| SIPA1L1 | CDR2L | 0.609062 | 4 | 0 | 3 |
| SIPA1L1 | RAD51 | 0.599276 | 3 | 0 | 3 |
| SIPA1L1 | ALDH8A1 | 0.594845 | 4 | 0 | 3 |
| SIPA1L1 | YEATS2 | 0.592489 | 3 | 0 | 3 |
| SIPA1L1 | TTLL5 | 0.583583 | 7 | 0 | 4 |
| SIPA1L1 | ZMYND15 | 0.583165 | 4 | 0 | 3 |
| SIPA1L1 | PGP | 0.582139 | 4 | 0 | 3 |
For details and further investigation, click here