| Full name: SKI like proto-oncogene | Alias Symbol: SNO|SnoN|SnoA | ||
| Type: protein-coding gene | Cytoband: 3q26.2 | ||
| Entrez ID: 6498 | HGNC ID: HGNC:10897 | Ensembl Gene: ENSG00000136603 | OMIM ID: 165340 |
| Drug and gene relationship at DGIdb | |||
Expression of SKIL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SKIL | 6498 | 225227_at | 0.7956 | 0.0997 | |
| GSE20347 | SKIL | 6498 | 217591_at | 0.6737 | 0.0004 | |
| GSE23400 | SKIL | 6498 | 217591_at | 0.4561 | 0.0000 | |
| GSE26886 | SKIL | 6498 | 225227_at | 1.0241 | 0.0010 | |
| GSE29001 | SKIL | 6498 | 217591_at | 0.9598 | 0.0083 | |
| GSE38129 | SKIL | 6498 | 217591_at | 0.6854 | 0.0547 | |
| GSE45670 | SKIL | 6498 | 225227_at | 0.5984 | 0.0036 | |
| GSE53622 | SKIL | 6498 | 14173 | 0.7685 | 0.0000 | |
| GSE53624 | SKIL | 6498 | 14173 | 0.6539 | 0.0000 | |
| GSE63941 | SKIL | 6498 | 225227_at | -0.3657 | 0.6781 | |
| GSE77861 | SKIL | 6498 | 225227_at | 0.6542 | 0.1168 | |
| GSE97050 | SKIL | 6498 | A_23_P351215 | 0.5577 | 0.2155 | |
| SRP007169 | SKIL | 6498 | RNAseq | 0.4104 | 0.1691 | |
| SRP008496 | SKIL | 6498 | RNAseq | 0.8742 | 0.0012 | |
| SRP064894 | SKIL | 6498 | RNAseq | 0.7159 | 0.0028 | |
| SRP133303 | SKIL | 6498 | RNAseq | 1.2898 | 0.0000 | |
| SRP159526 | SKIL | 6498 | RNAseq | 0.7855 | 0.0161 | |
| SRP193095 | SKIL | 6498 | RNAseq | 0.3802 | 0.0124 | |
| SRP219564 | SKIL | 6498 | RNAseq | 0.3561 | 0.2647 | |
| TCGA | SKIL | 6498 | RNAseq | 0.0631 | 0.4320 |
Upregulated datasets: 2; Downregulated datasets: 0.
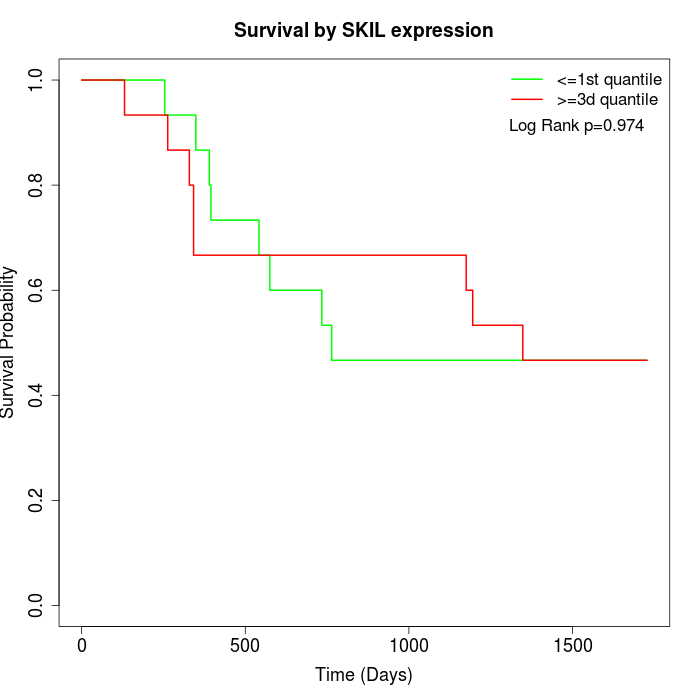
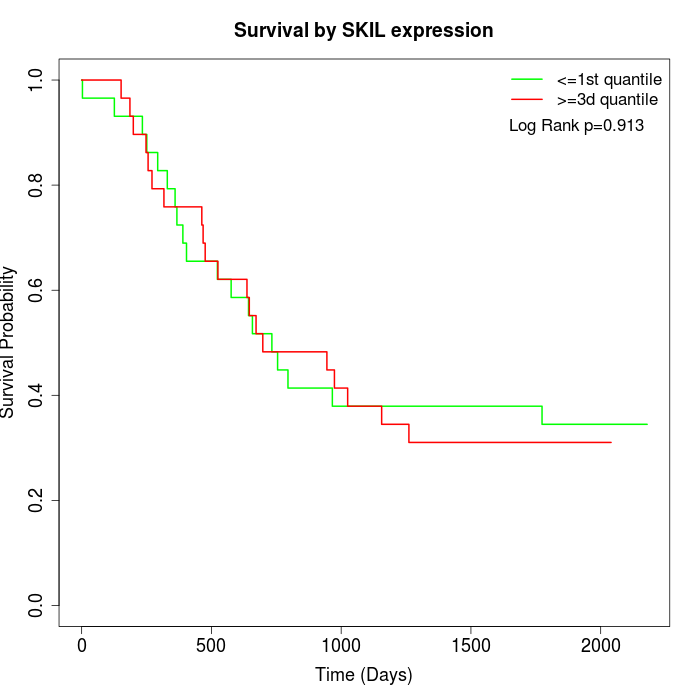
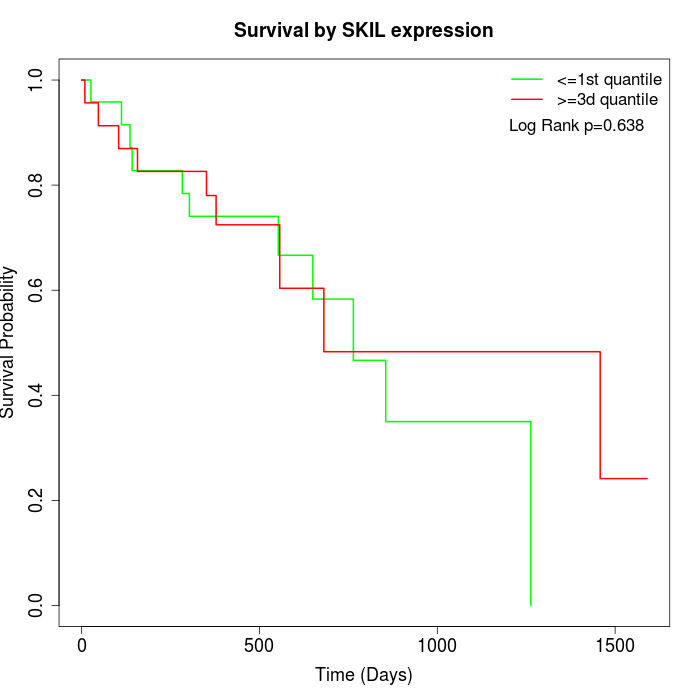
Survival by SKIL expression:
Note: Click image to view full size file.
Copy number change of SKIL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SKIL | 6498 | 22 | 0 | 8 | |
| GSE20123 | SKIL | 6498 | 22 | 0 | 8 | |
| GSE43470 | SKIL | 6498 | 24 | 0 | 19 | |
| GSE46452 | SKIL | 6498 | 19 | 1 | 39 | |
| GSE47630 | SKIL | 6498 | 27 | 2 | 11 | |
| GSE54993 | SKIL | 6498 | 1 | 14 | 55 | |
| GSE54994 | SKIL | 6498 | 41 | 0 | 12 | |
| GSE60625 | SKIL | 6498 | 0 | 6 | 5 | |
| GSE74703 | SKIL | 6498 | 20 | 0 | 16 | |
| GSE74704 | SKIL | 6498 | 14 | 0 | 6 | |
| TCGA | SKIL | 6498 | 80 | 0 | 16 |
Total number of gains: 270; Total number of losses: 23; Total Number of normals: 195.
Somatic mutations of SKIL:
Generating mutation plots.
Highly correlated genes for SKIL:
Showing top 20/594 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SKIL | TEP1 | 0.704455 | 3 | 0 | 3 |
| SKIL | BCAM | 0.680325 | 3 | 0 | 3 |
| SKIL | SLC28A3 | 0.677134 | 3 | 0 | 3 |
| SKIL | KIAA1614-AS1 | 0.660679 | 3 | 0 | 3 |
| SKIL | TTYH3 | 0.658509 | 7 | 0 | 5 |
| SKIL | KREMEN2 | 0.656 | 4 | 0 | 4 |
| SKIL | KIF26B | 0.652879 | 5 | 0 | 5 |
| SKIL | MAP3K7 | 0.64524 | 7 | 0 | 6 |
| SKIL | SCARB1 | 0.644156 | 5 | 0 | 5 |
| SKIL | TOMM40 | 0.641276 | 4 | 0 | 3 |
| SKIL | PDE7A | 0.640898 | 6 | 0 | 5 |
| SKIL | KIF3B | 0.640257 | 7 | 0 | 5 |
| SKIL | CTHRC1 | 0.640149 | 7 | 0 | 6 |
| SKIL | PRR5L | 0.637129 | 5 | 0 | 4 |
| SKIL | MTRR | 0.635687 | 3 | 0 | 3 |
| SKIL | FAF2 | 0.635651 | 4 | 0 | 4 |
| SKIL | CABLES2 | 0.634255 | 4 | 0 | 3 |
| SKIL | TP53I13 | 0.632724 | 4 | 0 | 4 |
| SKIL | LAMC2 | 0.631656 | 10 | 0 | 9 |
| SKIL | CD47 | 0.629936 | 3 | 0 | 3 |
For details and further investigation, click here