| Full name: laminin subunit gamma 2 | Alias Symbol: nicein-100kDa|kalinin-105kDa|BM600-100kDa | ||
| Type: protein-coding gene | Cytoband: 1q25.3 | ||
| Entrez ID: 3918 | HGNC ID: HGNC:6493 | Ensembl Gene: ENSG00000058085 | OMIM ID: 150292 |
| Drug and gene relationship at DGIdb | |||
LAMC2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa05200 | Pathways in cancer | |
| hsa05222 | Small cell lung cancer |
Expression of LAMC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAMC2 | 3918 | 202267_at | 2.6849 | 0.0086 | |
| GSE20347 | LAMC2 | 3918 | 202267_at | 2.4234 | 0.0000 | |
| GSE23400 | LAMC2 | 3918 | 202267_at | 1.8912 | 0.0000 | |
| GSE26886 | LAMC2 | 3918 | 202267_at | 4.0658 | 0.0000 | |
| GSE29001 | LAMC2 | 3918 | 202267_at | 2.5025 | 0.0001 | |
| GSE38129 | LAMC2 | 3918 | 202267_at | 2.8239 | 0.0000 | |
| GSE45670 | LAMC2 | 3918 | 202267_at | 3.1830 | 0.0001 | |
| GSE53622 | LAMC2 | 3918 | 27407 | 3.6482 | 0.0000 | |
| GSE53624 | LAMC2 | 3918 | 117153 | 3.1691 | 0.0000 | |
| GSE63941 | LAMC2 | 3918 | 202267_at | 2.7831 | 0.0049 | |
| GSE77861 | LAMC2 | 3918 | 202267_at | 2.3080 | 0.0151 | |
| GSE97050 | LAMC2 | 3918 | A_23_P160968 | 3.9143 | 0.0555 | |
| SRP007169 | LAMC2 | 3918 | RNAseq | 4.7155 | 0.0000 | |
| SRP008496 | LAMC2 | 3918 | RNAseq | 5.4186 | 0.0000 | |
| SRP064894 | LAMC2 | 3918 | RNAseq | 4.4181 | 0.0000 | |
| SRP133303 | LAMC2 | 3918 | RNAseq | 5.0638 | 0.0000 | |
| SRP159526 | LAMC2 | 3918 | RNAseq | 3.7772 | 0.0000 | |
| SRP193095 | LAMC2 | 3918 | RNAseq | 4.3057 | 0.0000 | |
| TCGA | LAMC2 | 3918 | RNAseq | 0.7716 | 0.0000 |
Upregulated datasets: 17; Downregulated datasets: 0.
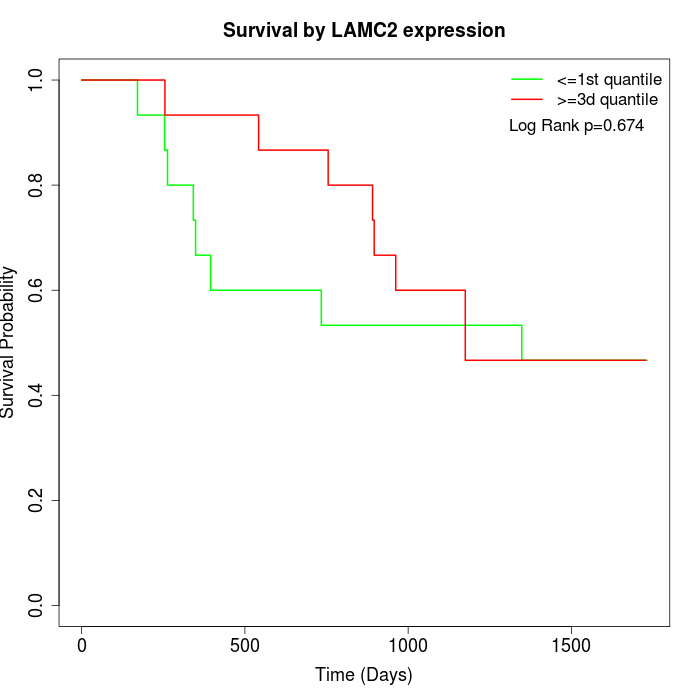
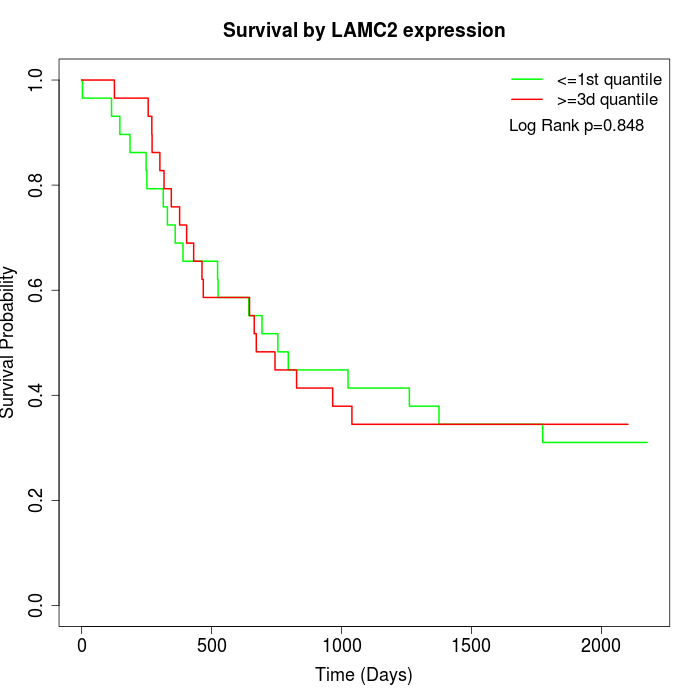
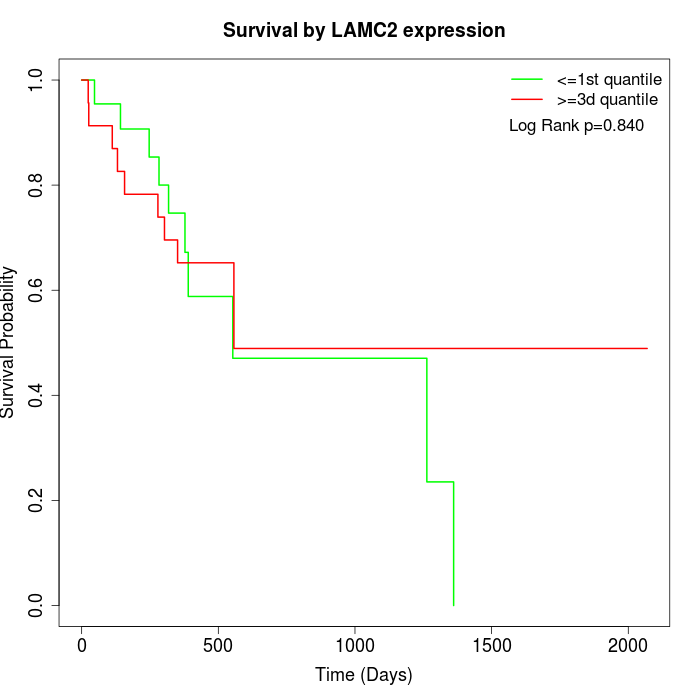
Survival by LAMC2 expression:
Note: Click image to view full size file.
Copy number change of LAMC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LAMC2 | 3918 | 12 | 0 | 18 | |
| GSE20123 | LAMC2 | 3918 | 12 | 0 | 18 | |
| GSE43470 | LAMC2 | 3918 | 7 | 1 | 35 | |
| GSE46452 | LAMC2 | 3918 | 3 | 1 | 55 | |
| GSE47630 | LAMC2 | 3918 | 14 | 0 | 26 | |
| GSE54993 | LAMC2 | 3918 | 0 | 6 | 64 | |
| GSE54994 | LAMC2 | 3918 | 15 | 0 | 38 | |
| GSE60625 | LAMC2 | 3918 | 0 | 0 | 11 | |
| GSE74703 | LAMC2 | 3918 | 7 | 1 | 28 | |
| GSE74704 | LAMC2 | 3918 | 5 | 0 | 15 | |
| TCGA | LAMC2 | 3918 | 40 | 3 | 53 |
Total number of gains: 115; Total number of losses: 12; Total Number of normals: 361.
Somatic mutations of LAMC2:
Generating mutation plots.
Highly correlated genes for LAMC2:
Showing top 20/1788 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAMC2 | CTHRC1 | 0.860583 | 7 | 0 | 7 |
| LAMC2 | PLAU | 0.835383 | 12 | 0 | 12 |
| LAMC2 | LAMA3 | 0.832638 | 12 | 0 | 12 |
| LAMC2 | LAMB3 | 0.818036 | 13 | 0 | 13 |
| LAMC2 | WDR54 | 0.810396 | 6 | 0 | 6 |
| LAMC2 | XPR1 | 0.809202 | 7 | 0 | 7 |
| LAMC2 | MMP1 | 0.806744 | 12 | 0 | 11 |
| LAMC2 | MFAP2 | 0.80128 | 12 | 0 | 11 |
| LAMC2 | ADAMTS2 | 0.798651 | 5 | 0 | 5 |
| LAMC2 | WDR66 | 0.796674 | 8 | 0 | 7 |
| LAMC2 | INHBA | 0.79608 | 11 | 0 | 10 |
| LAMC2 | TGFBI | 0.792604 | 12 | 0 | 12 |
| LAMC2 | CDH3 | 0.786114 | 11 | 0 | 11 |
| LAMC2 | PTHLH | 0.786012 | 12 | 0 | 11 |
| LAMC2 | ITGA6 | 0.785841 | 13 | 0 | 12 |
| LAMC2 | SPP1 | 0.783408 | 11 | 0 | 11 |
| LAMC2 | DCBLD1 | 0.780825 | 8 | 0 | 8 |
| LAMC2 | USB1 | 0.780531 | 10 | 0 | 10 |
| LAMC2 | COL7A1 | 0.775375 | 11 | 0 | 11 |
| LAMC2 | COLGALT1 | 0.775354 | 10 | 0 | 10 |
For details and further investigation, click here