| Full name: SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 | Alias Symbol: BAF170|Rsc8|CRACC2 | ||
| Type: protein-coding gene | Cytoband: 12q13.2 | ||
| Entrez ID: 6601 | HGNC ID: HGNC:11105 | Ensembl Gene: ENSG00000139613 | OMIM ID: 601734 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SMARCC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMARCC2 | 6601 | 201321_s_at | -0.1540 | 0.8140 | |
| GSE20347 | SMARCC2 | 6601 | 201321_s_at | -0.0215 | 0.9061 | |
| GSE23400 | SMARCC2 | 6601 | 201320_at | -0.0246 | 0.5834 | |
| GSE26886 | SMARCC2 | 6601 | 201320_at | -0.6319 | 0.0001 | |
| GSE29001 | SMARCC2 | 6601 | 201321_s_at | -0.1537 | 0.6981 | |
| GSE38129 | SMARCC2 | 6601 | 201321_s_at | -0.0865 | 0.5085 | |
| GSE45670 | SMARCC2 | 6601 | 201321_s_at | -0.2623 | 0.0086 | |
| GSE53622 | SMARCC2 | 6601 | 9516 | -0.3460 | 0.0000 | |
| GSE53624 | SMARCC2 | 6601 | 9516 | -0.1559 | 0.0074 | |
| GSE63941 | SMARCC2 | 6601 | 201321_s_at | 0.6372 | 0.0947 | |
| GSE77861 | SMARCC2 | 6601 | 201321_s_at | 0.2267 | 0.3459 | |
| GSE97050 | SMARCC2 | 6601 | A_23_P128073 | -0.4760 | 0.1490 | |
| SRP007169 | SMARCC2 | 6601 | RNAseq | 0.0168 | 0.9636 | |
| SRP008496 | SMARCC2 | 6601 | RNAseq | -0.1037 | 0.5934 | |
| SRP064894 | SMARCC2 | 6601 | RNAseq | -0.1141 | 0.3731 | |
| SRP133303 | SMARCC2 | 6601 | RNAseq | -0.0599 | 0.6702 | |
| SRP159526 | SMARCC2 | 6601 | RNAseq | -0.0815 | 0.6731 | |
| SRP193095 | SMARCC2 | 6601 | RNAseq | 0.0944 | 0.3926 | |
| SRP219564 | SMARCC2 | 6601 | RNAseq | 0.1245 | 0.7208 | |
| TCGA | SMARCC2 | 6601 | RNAseq | -0.0276 | 0.5089 |
Upregulated datasets: 0; Downregulated datasets: 0.
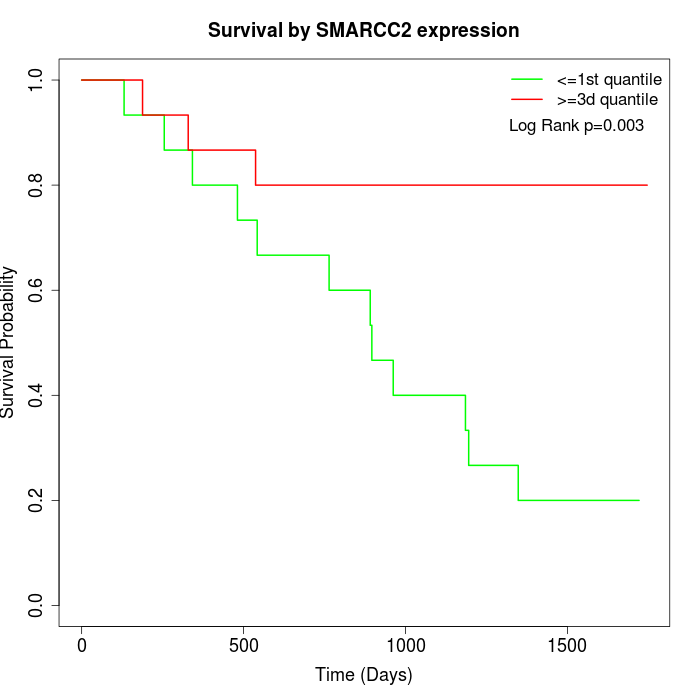
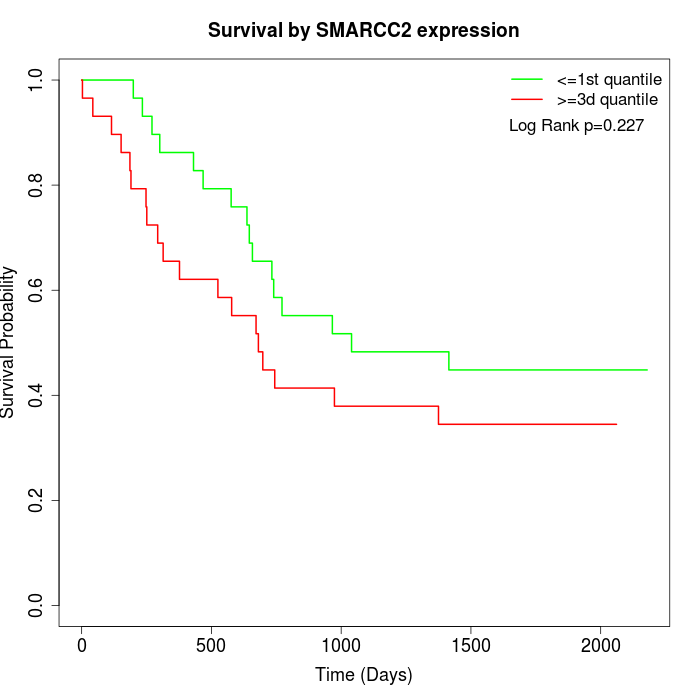
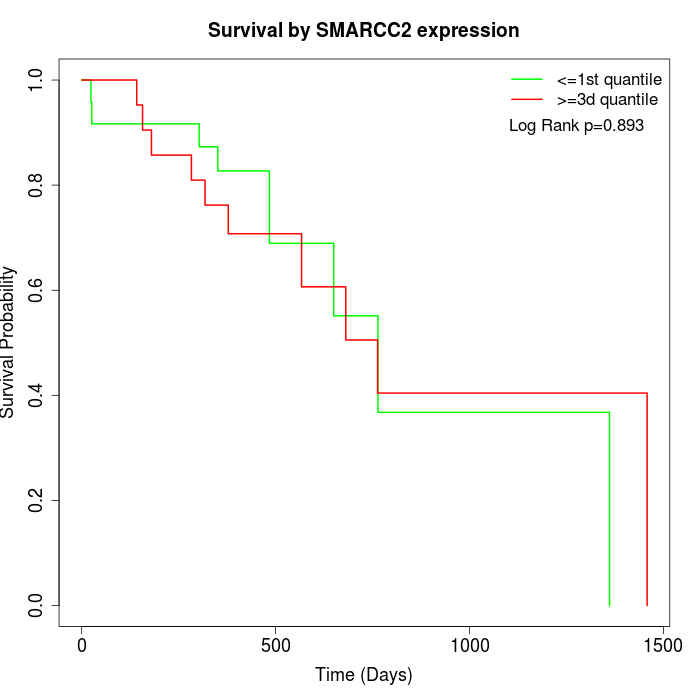
Survival by SMARCC2 expression:
Note: Click image to view full size file.
Copy number change of SMARCC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMARCC2 | 6601 | 7 | 2 | 21 | |
| GSE20123 | SMARCC2 | 6601 | 7 | 1 | 22 | |
| GSE43470 | SMARCC2 | 6601 | 4 | 0 | 39 | |
| GSE46452 | SMARCC2 | 6601 | 8 | 1 | 50 | |
| GSE47630 | SMARCC2 | 6601 | 10 | 2 | 28 | |
| GSE54993 | SMARCC2 | 6601 | 0 | 5 | 65 | |
| GSE54994 | SMARCC2 | 6601 | 4 | 1 | 48 | |
| GSE60625 | SMARCC2 | 6601 | 0 | 0 | 11 | |
| GSE74703 | SMARCC2 | 6601 | 4 | 0 | 32 | |
| GSE74704 | SMARCC2 | 6601 | 4 | 1 | 15 | |
| TCGA | SMARCC2 | 6601 | 14 | 10 | 72 |
Total number of gains: 62; Total number of losses: 23; Total Number of normals: 403.
Somatic mutations of SMARCC2:
Generating mutation plots.
Highly correlated genes for SMARCC2:
Showing top 20/327 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMARCC2 | C5orf51 | 0.825467 | 3 | 0 | 3 |
| SMARCC2 | IPO11 | 0.770772 | 3 | 0 | 3 |
| SMARCC2 | LCOR | 0.767359 | 3 | 0 | 3 |
| SMARCC2 | DNAJC21 | 0.763841 | 3 | 0 | 3 |
| SMARCC2 | AKAP17A | 0.742027 | 3 | 0 | 3 |
| SMARCC2 | KDM4A | 0.737494 | 3 | 0 | 3 |
| SMARCC2 | HSDL2 | 0.712217 | 3 | 0 | 3 |
| SMARCC2 | FAM172A | 0.708996 | 3 | 0 | 3 |
| SMARCC2 | MACROD2 | 0.69347 | 3 | 0 | 3 |
| SMARCC2 | TMEM80 | 0.691569 | 5 | 0 | 5 |
| SMARCC2 | ZFAND3 | 0.690666 | 4 | 0 | 3 |
| SMARCC2 | TRIM33 | 0.686519 | 5 | 0 | 4 |
| SMARCC2 | TJP1 | 0.678264 | 4 | 0 | 3 |
| SMARCC2 | SARM1 | 0.675618 | 4 | 0 | 4 |
| SMARCC2 | NR3C2 | 0.674909 | 4 | 0 | 4 |
| SMARCC2 | C11orf68 | 0.674387 | 4 | 0 | 3 |
| SMARCC2 | KCNAB1 | 0.673503 | 5 | 0 | 4 |
| SMARCC2 | ZNF346 | 0.670709 | 4 | 0 | 4 |
| SMARCC2 | HECTD3 | 0.669983 | 4 | 0 | 3 |
| SMARCC2 | CCDC159 | 0.669156 | 4 | 0 | 3 |
For details and further investigation, click here