| Full name: tight junction protein 1 | Alias Symbol: ZO-1|MGC133289|DKFZp686M05161 | ||
| Type: protein-coding gene | Cytoband: 15q13.1 | ||
| Entrez ID: 7082 | HGNC ID: HGNC:11827 | Ensembl Gene: ENSG00000104067 | OMIM ID: 601009 |
| Drug and gene relationship at DGIdb | |||
TJP1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04520 | Adherens junction | |
| hsa04530 | Tight junction | |
| hsa04540 | Gap junction | |
| hsa05132 | Salmonella infection |
Expression of TJP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TJP1 | 7082 | 202011_at | -0.9091 | 0.1693 | |
| GSE20347 | TJP1 | 7082 | 202011_at | -1.2912 | 0.0000 | |
| GSE23400 | TJP1 | 7082 | 202011_at | -1.0667 | 0.0000 | |
| GSE26886 | TJP1 | 7082 | 202011_at | -1.2571 | 0.0000 | |
| GSE29001 | TJP1 | 7082 | 202011_at | -0.9210 | 0.0003 | |
| GSE38129 | TJP1 | 7082 | 202011_at | -1.0786 | 0.0000 | |
| GSE45670 | TJP1 | 7082 | 202011_at | -0.5951 | 0.0013 | |
| GSE53622 | TJP1 | 7082 | 57273 | -1.1264 | 0.0000 | |
| GSE53624 | TJP1 | 7082 | 57273 | -1.2335 | 0.0000 | |
| GSE63941 | TJP1 | 7082 | 202011_at | -0.2575 | 0.5714 | |
| GSE77861 | TJP1 | 7082 | 202011_at | -0.7748 | 0.0081 | |
| GSE97050 | TJP1 | 7082 | A_24_P193435 | -0.8442 | 0.0631 | |
| SRP007169 | TJP1 | 7082 | RNAseq | -1.0750 | 0.0194 | |
| SRP008496 | TJP1 | 7082 | RNAseq | -1.1485 | 0.0001 | |
| SRP064894 | TJP1 | 7082 | RNAseq | -1.9283 | 0.0000 | |
| SRP133303 | TJP1 | 7082 | RNAseq | -0.9767 | 0.0000 | |
| SRP159526 | TJP1 | 7082 | RNAseq | -1.6831 | 0.0000 | |
| SRP193095 | TJP1 | 7082 | RNAseq | -1.2588 | 0.0000 | |
| SRP219564 | TJP1 | 7082 | RNAseq | -1.7475 | 0.0003 | |
| TCGA | TJP1 | 7082 | RNAseq | -0.0999 | 0.0677 |
Upregulated datasets: 0; Downregulated datasets: 12.
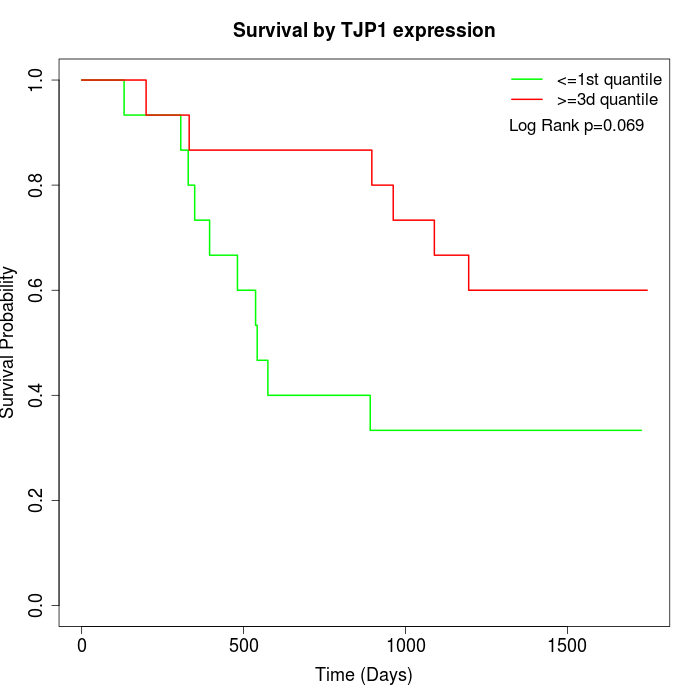
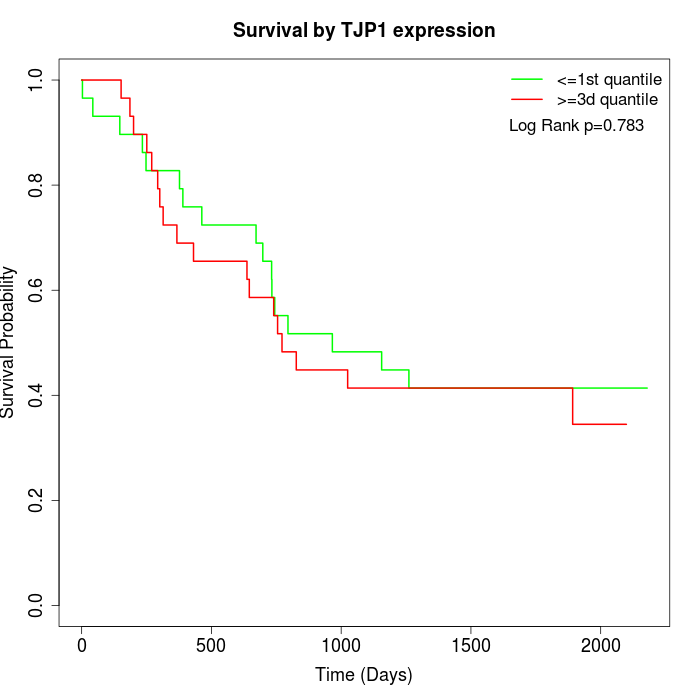
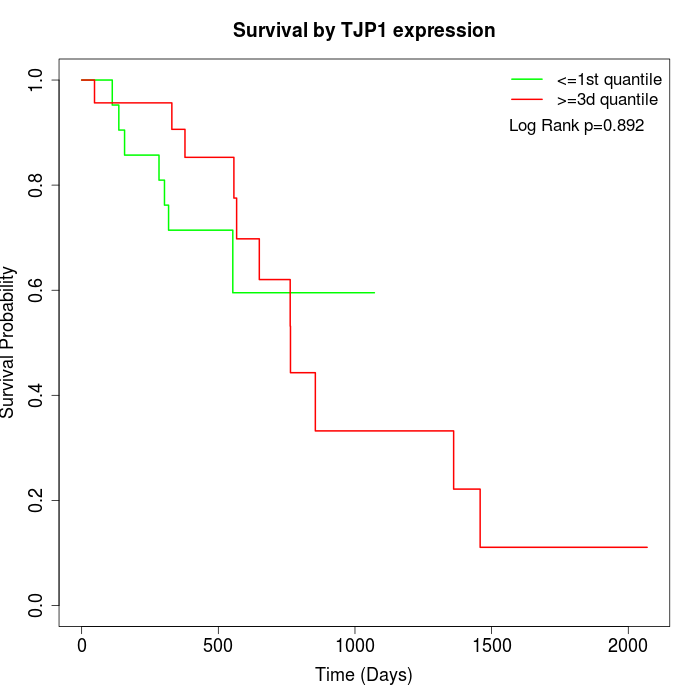
Survival by TJP1 expression:
Note: Click image to view full size file.
Copy number change of TJP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TJP1 | 7082 | 2 | 7 | 21 | |
| GSE20123 | TJP1 | 7082 | 2 | 7 | 21 | |
| GSE43470 | TJP1 | 7082 | 4 | 4 | 35 | |
| GSE46452 | TJP1 | 7082 | 3 | 7 | 49 | |
| GSE47630 | TJP1 | 7082 | 9 | 10 | 21 | |
| GSE54993 | TJP1 | 7082 | 4 | 6 | 60 | |
| GSE54994 | TJP1 | 7082 | 6 | 7 | 40 | |
| GSE60625 | TJP1 | 7082 | 4 | 0 | 7 | |
| GSE74703 | TJP1 | 7082 | 4 | 3 | 29 | |
| GSE74704 | TJP1 | 7082 | 1 | 6 | 13 | |
| TCGA | TJP1 | 7082 | 16 | 16 | 64 |
Total number of gains: 55; Total number of losses: 73; Total Number of normals: 360.
Somatic mutations of TJP1:
Generating mutation plots.
Highly correlated genes for TJP1:
Showing top 20/1783 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TJP1 | RAB11A | 0.859416 | 11 | 0 | 11 |
| TJP1 | TAB3 | 0.844176 | 7 | 0 | 7 |
| TJP1 | SRP68 | 0.831424 | 3 | 0 | 3 |
| TJP1 | IMPACT | 0.830981 | 3 | 0 | 3 |
| TJP1 | SASH1 | 0.821288 | 11 | 0 | 11 |
| TJP1 | CHP1 | 0.821192 | 10 | 0 | 10 |
| TJP1 | FRMD4B | 0.819381 | 11 | 0 | 11 |
| TJP1 | RNF141 | 0.81537 | 11 | 0 | 11 |
| TJP1 | CPEB3 | 0.809597 | 10 | 0 | 10 |
| TJP1 | CCNYL1 | 0.807594 | 5 | 0 | 5 |
| TJP1 | GDPD3 | 0.806614 | 10 | 0 | 10 |
| TJP1 | EVPL | 0.802938 | 10 | 0 | 10 |
| TJP1 | MAL | 0.801149 | 10 | 0 | 10 |
| TJP1 | MGLL | 0.799153 | 11 | 0 | 11 |
| TJP1 | SORT1 | 0.796943 | 11 | 0 | 11 |
| TJP1 | CYSRT1 | 0.795845 | 6 | 0 | 6 |
| TJP1 | ARHGEF10L | 0.794556 | 11 | 0 | 11 |
| TJP1 | EPS8L1 | 0.793836 | 10 | 0 | 10 |
| TJP1 | PDLIM2 | 0.793336 | 9 | 0 | 9 |
| TJP1 | EMP1 | 0.793219 | 11 | 0 | 10 |
For details and further investigation, click here