| Full name: Sp1 transcription factor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.13 | ||
| Entrez ID: 6667 | HGNC ID: HGNC:11205 | Ensembl Gene: ENSG00000185591 | OMIM ID: 189906 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
SP1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04350 | TGF-beta signaling pathway | |
| hsa04915 | Estrogen signaling pathway | |
| hsa05231 | Choline metabolism in cancer |
Expression of SP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SP1 | 6667 | 224754_at | -0.2235 | 0.6689 | |
| GSE20347 | SP1 | 6667 | 214732_at | -0.1377 | 0.0762 | |
| GSE23400 | SP1 | 6667 | 214732_at | -0.2472 | 0.0000 | |
| GSE26886 | SP1 | 6667 | 224754_at | -0.6508 | 0.0057 | |
| GSE29001 | SP1 | 6667 | 214732_at | -0.1912 | 0.2928 | |
| GSE38129 | SP1 | 6667 | 214732_at | -0.1510 | 0.0917 | |
| GSE45670 | SP1 | 6667 | 224760_at | -0.2455 | 0.0929 | |
| GSE53622 | SP1 | 6667 | 106717 | -1.1923 | 0.0000 | |
| GSE53624 | SP1 | 6667 | 106717 | -1.8325 | 0.0000 | |
| GSE63941 | SP1 | 6667 | 224754_at | 0.9289 | 0.0389 | |
| GSE77861 | SP1 | 6667 | 224754_at | 0.0567 | 0.7605 | |
| GSE97050 | SP1 | 6667 | A_33_P3330283 | -0.5348 | 0.1133 | |
| SRP007169 | SP1 | 6667 | RNAseq | -0.6156 | 0.0688 | |
| SRP008496 | SP1 | 6667 | RNAseq | -0.6698 | 0.0000 | |
| SRP064894 | SP1 | 6667 | RNAseq | -0.5449 | 0.0005 | |
| SRP133303 | SP1 | 6667 | RNAseq | -0.2847 | 0.0032 | |
| SRP159526 | SP1 | 6667 | RNAseq | -0.6740 | 0.0055 | |
| SRP193095 | SP1 | 6667 | RNAseq | -0.3659 | 0.0000 | |
| SRP219564 | SP1 | 6667 | RNAseq | -0.7945 | 0.0566 | |
| TCGA | SP1 | 6667 | RNAseq | 0.0096 | 0.8112 |
Upregulated datasets: 0; Downregulated datasets: 2.
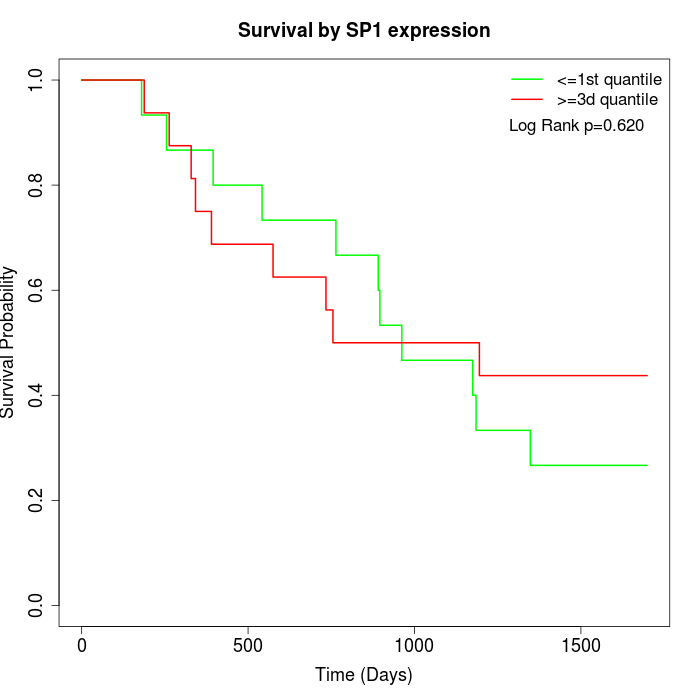
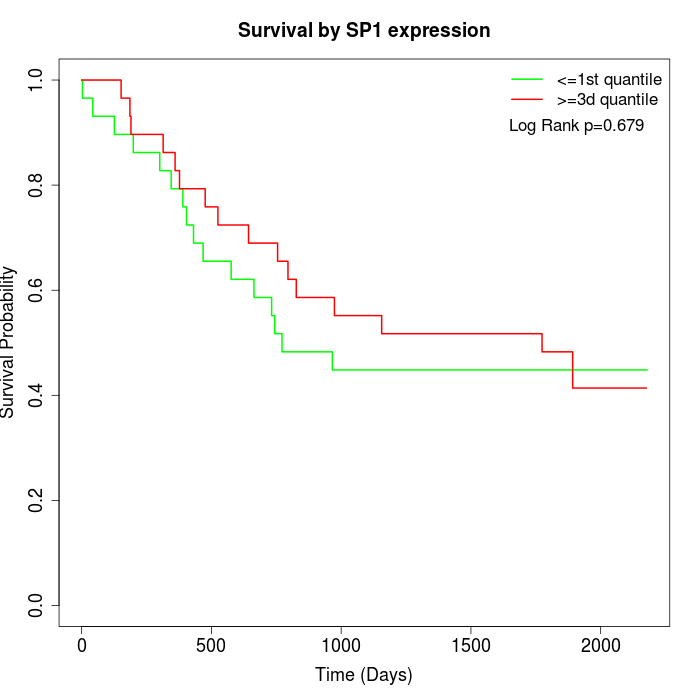
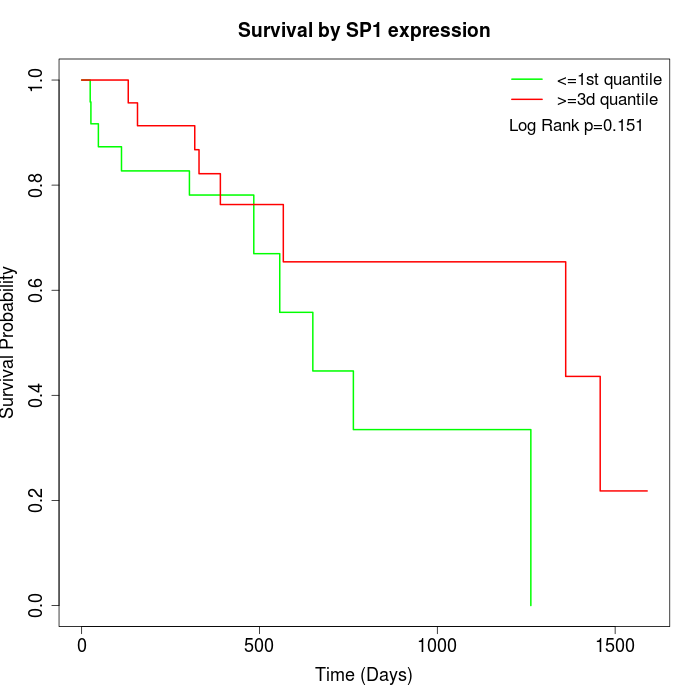
Survival by SP1 expression:
Note: Click image to view full size file.
Copy number change of SP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SP1 | 6667 | 7 | 1 | 22 | |
| GSE20123 | SP1 | 6667 | 7 | 1 | 22 | |
| GSE43470 | SP1 | 6667 | 3 | 0 | 40 | |
| GSE46452 | SP1 | 6667 | 7 | 1 | 51 | |
| GSE47630 | SP1 | 6667 | 10 | 2 | 28 | |
| GSE54993 | SP1 | 6667 | 0 | 5 | 65 | |
| GSE54994 | SP1 | 6667 | 4 | 1 | 48 | |
| GSE60625 | SP1 | 6667 | 0 | 0 | 11 | |
| GSE74703 | SP1 | 6667 | 3 | 0 | 33 | |
| GSE74704 | SP1 | 6667 | 5 | 1 | 14 | |
| TCGA | SP1 | 6667 | 14 | 11 | 71 |
Total number of gains: 60; Total number of losses: 23; Total Number of normals: 405.
Somatic mutations of SP1:
Generating mutation plots.
Highly correlated genes for SP1:
Showing top 20/818 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SP1 | ING4 | 0.844582 | 3 | 0 | 3 |
| SP1 | CASC3 | 0.83704 | 3 | 0 | 3 |
| SP1 | NTNG1 | 0.828309 | 3 | 0 | 3 |
| SP1 | TMEM218 | 0.817139 | 3 | 0 | 3 |
| SP1 | NUFIP2 | 0.783674 | 3 | 0 | 3 |
| SP1 | ATRNL1 | 0.773511 | 4 | 0 | 4 |
| SP1 | CA13 | 0.772088 | 3 | 0 | 3 |
| SP1 | TXNDC16 | 0.76853 | 3 | 0 | 3 |
| SP1 | SLC10A5 | 0.759622 | 3 | 0 | 3 |
| SP1 | SHPRH | 0.758253 | 3 | 0 | 3 |
| SP1 | HOXA3 | 0.753144 | 3 | 0 | 3 |
| SP1 | ULK3 | 0.743021 | 5 | 0 | 5 |
| SP1 | ERP27 | 0.741373 | 3 | 0 | 3 |
| SP1 | ATXN7L3B | 0.740012 | 4 | 0 | 3 |
| SP1 | ZNF780B | 0.739771 | 3 | 0 | 3 |
| SP1 | ARFGAP2 | 0.739599 | 4 | 0 | 3 |
| SP1 | A2ML1 | 0.738379 | 4 | 0 | 4 |
| SP1 | ARL6 | 0.738271 | 3 | 0 | 3 |
| SP1 | COPS7A | 0.737231 | 3 | 0 | 3 |
| SP1 | MRPS36 | 0.733929 | 4 | 0 | 4 |
For details and further investigation, click here