| Full name: signal peptidase complex subunit 2 | Alias Symbol: KIAA0102 | ||
| Type: protein-coding gene | Cytoband: 11q13.4 | ||
| Entrez ID: 9789 | HGNC ID: HGNC:28962 | Ensembl Gene: ENSG00000118363 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SPCS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPCS2 | 9789 | 201240_s_at | 0.2677 | 0.6789 | |
| GSE20347 | SPCS2 | 9789 | 201240_s_at | 0.1474 | 0.3344 | |
| GSE23400 | SPCS2 | 9789 | 201240_s_at | 0.5943 | 0.0000 | |
| GSE26886 | SPCS2 | 9789 | 201240_s_at | -0.3797 | 0.0420 | |
| GSE29001 | SPCS2 | 9789 | 201240_s_at | 0.1274 | 0.6346 | |
| GSE38129 | SPCS2 | 9789 | 201240_s_at | 0.1785 | 0.1228 | |
| GSE45670 | SPCS2 | 9789 | 201240_s_at | 0.2233 | 0.1471 | |
| GSE53622 | SPCS2 | 9789 | 9661 | 0.1256 | 0.0937 | |
| GSE53624 | SPCS2 | 9789 | 9661 | 0.1627 | 0.0085 | |
| GSE63941 | SPCS2 | 9789 | 201240_s_at | -0.0280 | 0.9539 | |
| GSE77861 | SPCS2 | 9789 | 201240_s_at | 0.0101 | 0.9794 | |
| GSE97050 | SPCS2 | 9789 | A_23_P2143 | 0.2295 | 0.5215 | |
| SRP007169 | SPCS2 | 9789 | RNAseq | 0.1837 | 0.6585 | |
| SRP008496 | SPCS2 | 9789 | RNAseq | 0.3583 | 0.2033 | |
| SRP064894 | SPCS2 | 9789 | RNAseq | 0.2484 | 0.1838 | |
| SRP133303 | SPCS2 | 9789 | RNAseq | 0.0109 | 0.9420 | |
| SRP159526 | SPCS2 | 9789 | RNAseq | -0.1658 | 0.5774 | |
| SRP193095 | SPCS2 | 9789 | RNAseq | -0.2549 | 0.0921 | |
| SRP219564 | SPCS2 | 9789 | RNAseq | 0.4782 | 0.2806 | |
| TCGA | SPCS2 | 9789 | RNAseq | 0.0361 | 0.6357 |
Upregulated datasets: 0; Downregulated datasets: 0.
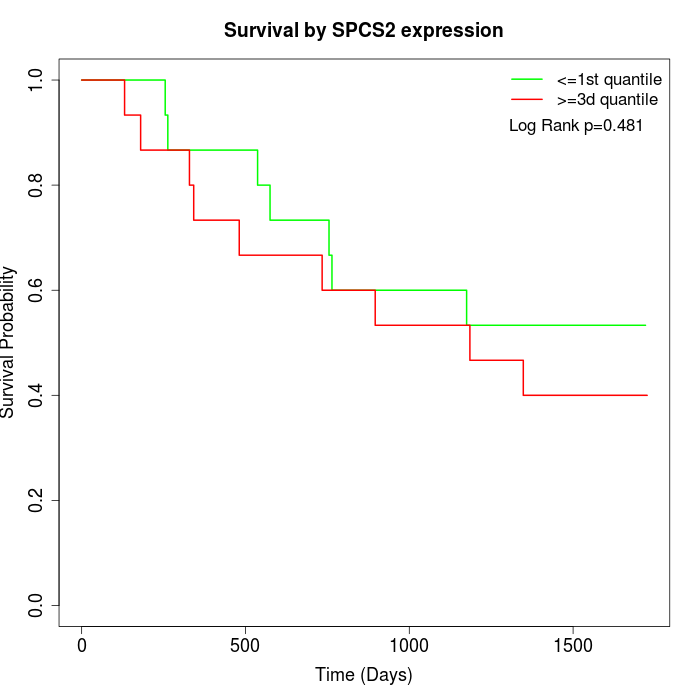
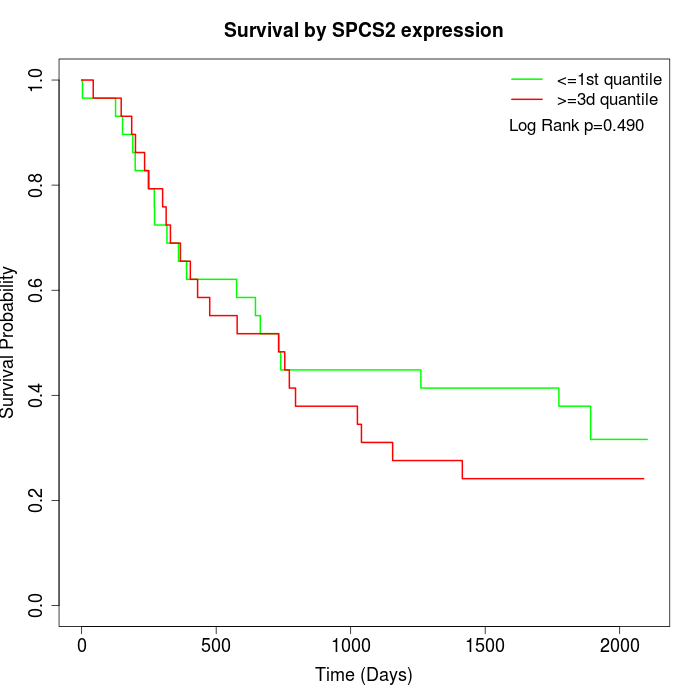
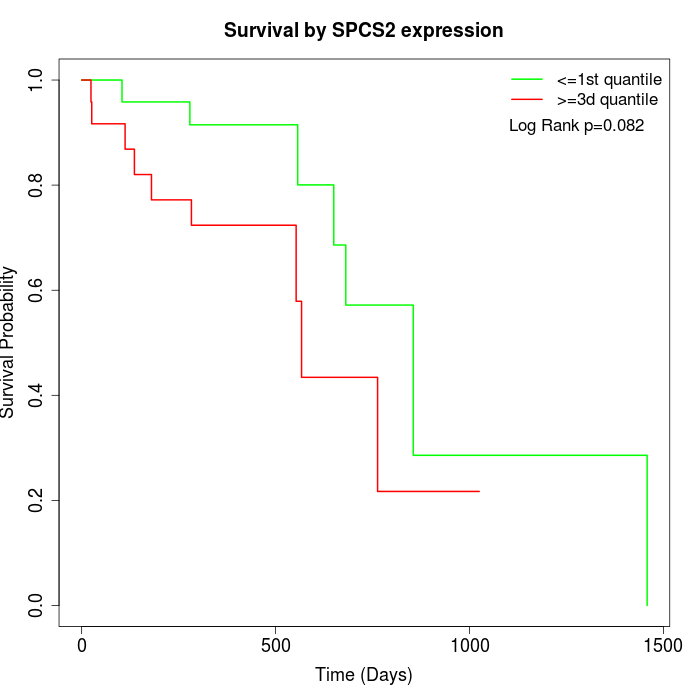
Survival by SPCS2 expression:
Note: Click image to view full size file.
Copy number change of SPCS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPCS2 | 9789 | 6 | 8 | 16 | |
| GSE20123 | SPCS2 | 9789 | 6 | 7 | 17 | |
| GSE43470 | SPCS2 | 9789 | 5 | 4 | 34 | |
| GSE46452 | SPCS2 | 9789 | 7 | 22 | 30 | |
| GSE47630 | SPCS2 | 9789 | 6 | 14 | 20 | |
| GSE54993 | SPCS2 | 9789 | 8 | 3 | 59 | |
| GSE54994 | SPCS2 | 9789 | 8 | 15 | 30 | |
| GSE60625 | SPCS2 | 9789 | 4 | 3 | 4 | |
| GSE74703 | SPCS2 | 9789 | 4 | 3 | 29 | |
| GSE74704 | SPCS2 | 9789 | 4 | 3 | 13 | |
| TCGA | SPCS2 | 9789 | 22 | 33 | 41 |
Total number of gains: 80; Total number of losses: 115; Total Number of normals: 293.
Somatic mutations of SPCS2:
Generating mutation plots.
Highly correlated genes for SPCS2:
Showing top 20/302 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPCS2 | PTS | 0.774598 | 3 | 0 | 3 |
| SPCS2 | TRAPPC6B | 0.766135 | 3 | 0 | 3 |
| SPCS2 | ANKS6 | 0.76539 | 3 | 0 | 3 |
| SPCS2 | FH | 0.76097 | 3 | 0 | 3 |
| SPCS2 | NCKAP1 | 0.760333 | 3 | 0 | 3 |
| SPCS2 | EIF4G1 | 0.760121 | 3 | 0 | 3 |
| SPCS2 | B3GALNT2 | 0.746244 | 3 | 0 | 3 |
| SPCS2 | RASA2 | 0.744261 | 3 | 0 | 3 |
| SPCS2 | EIF3C | 0.738629 | 3 | 0 | 3 |
| SPCS2 | CMTM6 | 0.733681 | 4 | 0 | 4 |
| SPCS2 | ZNF300 | 0.727648 | 3 | 0 | 3 |
| SPCS2 | CEBPZ | 0.727561 | 3 | 0 | 3 |
| SPCS2 | RBM23 | 0.727172 | 4 | 0 | 3 |
| SPCS2 | NDUFA9 | 0.725136 | 3 | 0 | 3 |
| SPCS2 | VANGL1 | 0.724907 | 3 | 0 | 3 |
| SPCS2 | GOLIM4 | 0.72474 | 3 | 0 | 3 |
| SPCS2 | PCTP | 0.724268 | 3 | 0 | 3 |
| SPCS2 | NDUFA10 | 0.72216 | 4 | 0 | 4 |
| SPCS2 | NDUFS3 | 0.721008 | 3 | 0 | 3 |
| SPCS2 | COASY | 0.717607 | 3 | 0 | 3 |
For details and further investigation, click here