| Full name: squalene epoxidase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 8q24.13 | ||
| Entrez ID: 6713 | HGNC ID: HGNC:11279 | Ensembl Gene: ENSG00000104549 | OMIM ID: 602019 |
| Drug and gene relationship at DGIdb | |||
Expression of SQLE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SQLE | 6713 | 209218_at | 0.7276 | 0.4848 | |
| GSE20347 | SQLE | 6713 | 209218_at | 0.3825 | 0.1058 | |
| GSE23400 | SQLE | 6713 | 209218_at | 0.5010 | 0.0008 | |
| GSE26886 | SQLE | 6713 | 209218_at | -0.7915 | 0.0061 | |
| GSE29001 | SQLE | 6713 | 209218_at | 0.0566 | 0.9047 | |
| GSE38129 | SQLE | 6713 | 209218_at | 0.9939 | 0.0037 | |
| GSE45670 | SQLE | 6713 | 209218_at | 0.5903 | 0.0805 | |
| GSE53622 | SQLE | 6713 | 16528 | 0.6764 | 0.0001 | |
| GSE53624 | SQLE | 6713 | 16528 | 0.5215 | 0.0000 | |
| GSE63941 | SQLE | 6713 | 209218_at | -0.3584 | 0.6874 | |
| GSE77861 | SQLE | 6713 | 209218_at | 0.1196 | 0.6903 | |
| GSE97050 | SQLE | 6713 | A_23_P146284 | 0.9877 | 0.2110 | |
| SRP007169 | SQLE | 6713 | RNAseq | -0.2078 | 0.6324 | |
| SRP008496 | SQLE | 6713 | RNAseq | -0.3320 | 0.2135 | |
| SRP064894 | SQLE | 6713 | RNAseq | 0.0994 | 0.7551 | |
| SRP133303 | SQLE | 6713 | RNAseq | 0.9938 | 0.0105 | |
| SRP159526 | SQLE | 6713 | RNAseq | 0.1397 | 0.7265 | |
| SRP193095 | SQLE | 6713 | RNAseq | 0.5254 | 0.0178 | |
| SRP219564 | SQLE | 6713 | RNAseq | -0.0346 | 0.9659 | |
| TCGA | SQLE | 6713 | RNAseq | 0.5602 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
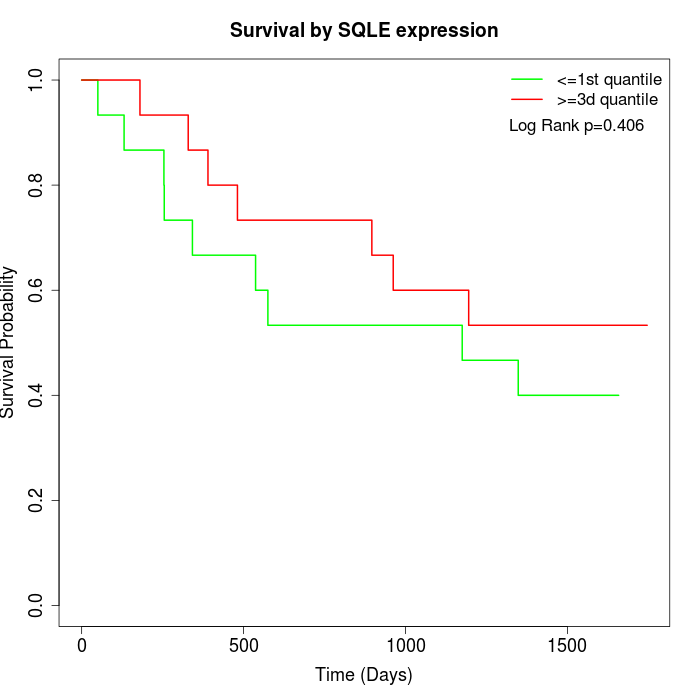
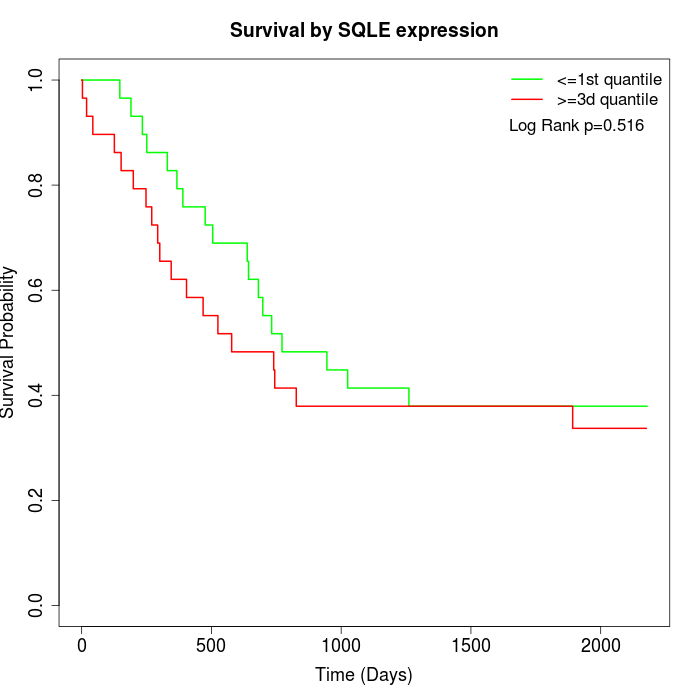
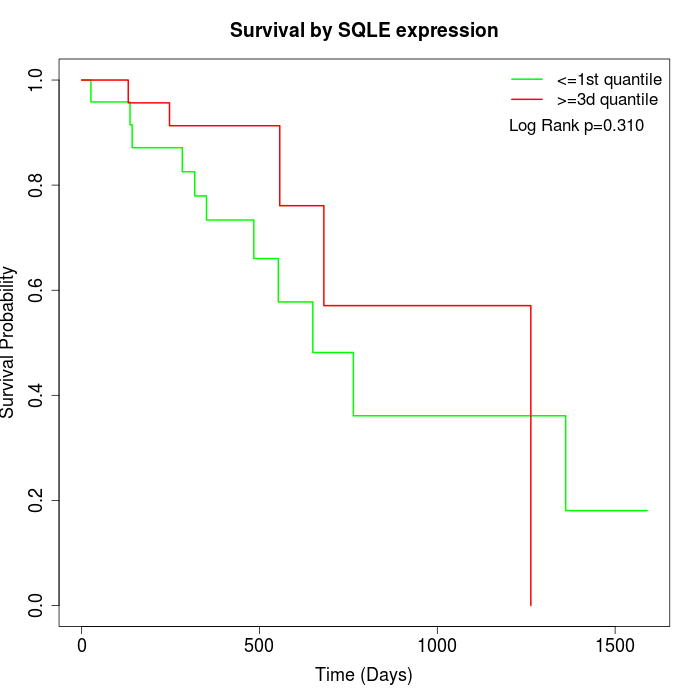
Survival by SQLE expression:
Note: Click image to view full size file.
Copy number change of SQLE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SQLE | 6713 | 20 | 0 | 10 | |
| GSE20123 | SQLE | 6713 | 21 | 0 | 9 | |
| GSE43470 | SQLE | 6713 | 22 | 0 | 21 | |
| GSE46452 | SQLE | 6713 | 28 | 0 | 31 | |
| GSE47630 | SQLE | 6713 | 24 | 0 | 16 | |
| GSE54993 | SQLE | 6713 | 0 | 21 | 49 | |
| GSE54994 | SQLE | 6713 | 37 | 1 | 15 | |
| GSE60625 | SQLE | 6713 | 0 | 4 | 7 | |
| GSE74703 | SQLE | 6713 | 19 | 0 | 17 | |
| GSE74704 | SQLE | 6713 | 13 | 0 | 7 | |
| TCGA | SQLE | 6713 | 68 | 1 | 27 |
Total number of gains: 252; Total number of losses: 27; Total Number of normals: 209.
Somatic mutations of SQLE:
Generating mutation plots.
Highly correlated genes for SQLE:
Showing top 20/343 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SQLE | TLCD1 | 0.766102 | 3 | 0 | 3 |
| SQLE | BIN3 | 0.690531 | 3 | 0 | 3 |
| SQLE | CENPK | 0.689567 | 3 | 0 | 3 |
| SQLE | FERMT1 | 0.681154 | 8 | 0 | 6 |
| SQLE | WFDC5 | 0.680095 | 4 | 0 | 4 |
| SQLE | PKP1 | 0.670567 | 6 | 0 | 5 |
| SQLE | VDR | 0.669693 | 5 | 0 | 4 |
| SQLE | CDH1 | 0.668189 | 6 | 0 | 6 |
| SQLE | SDC1 | 0.667093 | 6 | 0 | 4 |
| SQLE | AP1G1 | 0.666671 | 3 | 0 | 3 |
| SQLE | AP1S3 | 0.655929 | 4 | 0 | 4 |
| SQLE | CENPU | 0.655167 | 3 | 0 | 3 |
| SQLE | KCNK1 | 0.647256 | 7 | 0 | 5 |
| SQLE | FAM83B | 0.645204 | 5 | 0 | 3 |
| SQLE | FAM83H | 0.644515 | 6 | 0 | 4 |
| SQLE | ACTR2 | 0.642427 | 4 | 0 | 4 |
| SQLE | GGCT | 0.641692 | 7 | 0 | 6 |
| SQLE | AMMECR1 | 0.641119 | 5 | 0 | 4 |
| SQLE | FDPS | 0.64056 | 7 | 0 | 6 |
| SQLE | PIGW | 0.639863 | 4 | 0 | 3 |
For details and further investigation, click here