| Full name: gamma-glutamylcyclotransferase | Alias Symbol: MGC3077|CRF21|Ggc | ||
| Type: protein-coding gene | Cytoband: 7p14.3 | ||
| Entrez ID: 79017 | HGNC ID: HGNC:21705 | Ensembl Gene: ENSG00000006625 | OMIM ID: 137170 |
| Drug and gene relationship at DGIdb | |||
Expression of GGCT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GGCT | 79017 | 215380_s_at | 0.1992 | 0.6908 | |
| GSE20347 | GGCT | 79017 | 215380_s_at | 0.3625 | 0.0027 | |
| GSE23400 | GGCT | 79017 | 215380_s_at | 0.4063 | 0.0000 | |
| GSE26886 | GGCT | 79017 | 215380_s_at | -0.4248 | 0.0073 | |
| GSE29001 | GGCT | 79017 | 215380_s_at | 0.2714 | 0.1808 | |
| GSE38129 | GGCT | 79017 | 215380_s_at | 0.8017 | 0.0000 | |
| GSE45670 | GGCT | 79017 | 215380_s_at | 0.3123 | 0.0359 | |
| GSE53622 | GGCT | 79017 | 9188 | 0.0739 | 0.4001 | |
| GSE53624 | GGCT | 79017 | 9188 | 0.2887 | 0.0000 | |
| GSE63941 | GGCT | 79017 | 215380_s_at | 1.3787 | 0.0013 | |
| GSE77861 | GGCT | 79017 | 215380_s_at | 0.1649 | 0.6607 | |
| GSE97050 | GGCT | 79017 | A_23_P42695 | 0.4032 | 0.4400 | |
| SRP007169 | GGCT | 79017 | RNAseq | -0.3960 | 0.2672 | |
| SRP008496 | GGCT | 79017 | RNAseq | -0.1090 | 0.7099 | |
| SRP064894 | GGCT | 79017 | RNAseq | 0.2034 | 0.2785 | |
| SRP133303 | GGCT | 79017 | RNAseq | 0.1758 | 0.3892 | |
| SRP159526 | GGCT | 79017 | RNAseq | 0.3015 | 0.1286 | |
| SRP193095 | GGCT | 79017 | RNAseq | -0.3447 | 0.0733 | |
| SRP219564 | GGCT | 79017 | RNAseq | -0.1353 | 0.6995 | |
| TCGA | GGCT | 79017 | RNAseq | 0.3475 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
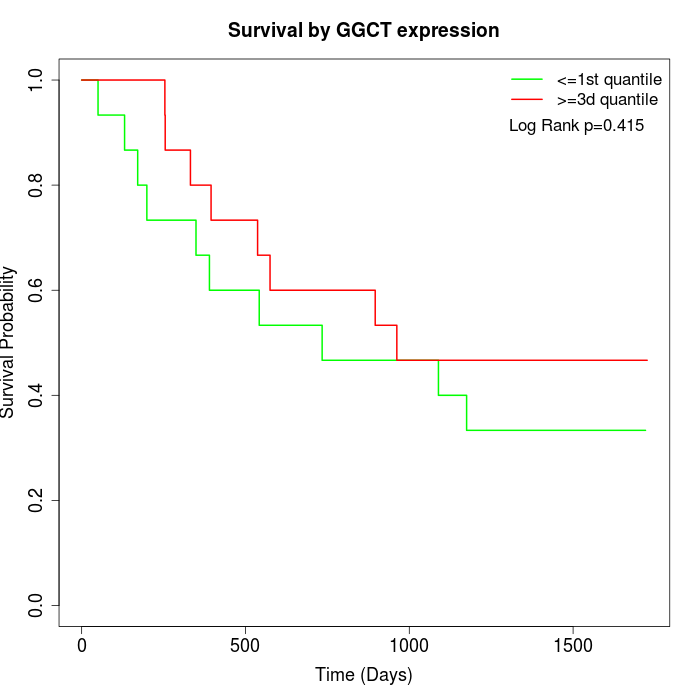
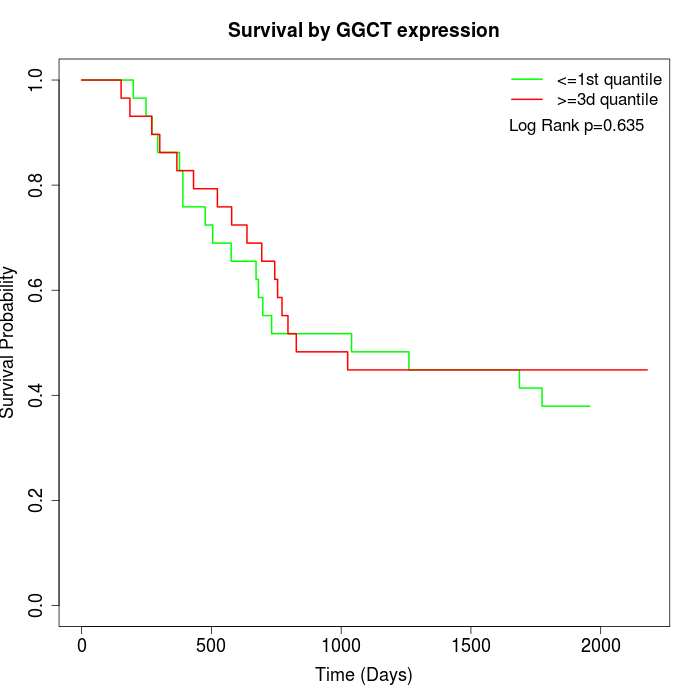
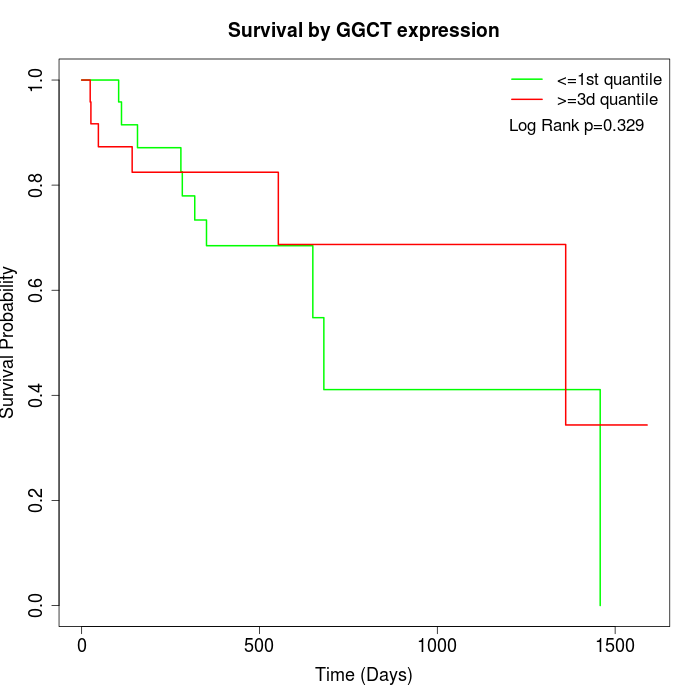
Survival by GGCT expression:
Note: Click image to view full size file.
Copy number change of GGCT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GGCT | 79017 | 15 | 0 | 15 | |
| GSE20123 | GGCT | 79017 | 15 | 0 | 15 | |
| GSE43470 | GGCT | 79017 | 4 | 0 | 39 | |
| GSE46452 | GGCT | 79017 | 11 | 1 | 47 | |
| GSE47630 | GGCT | 79017 | 8 | 1 | 31 | |
| GSE54993 | GGCT | 79017 | 0 | 6 | 64 | |
| GSE54994 | GGCT | 79017 | 19 | 3 | 31 | |
| GSE60625 | GGCT | 79017 | 0 | 0 | 11 | |
| GSE74703 | GGCT | 79017 | 4 | 0 | 32 | |
| GSE74704 | GGCT | 79017 | 9 | 0 | 11 | |
| TCGA | GGCT | 79017 | 55 | 7 | 34 |
Total number of gains: 140; Total number of losses: 18; Total Number of normals: 330.
Somatic mutations of GGCT:
Generating mutation plots.
Highly correlated genes for GGCT:
Showing top 20/392 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GGCT | FAM83B | 0.762624 | 4 | 0 | 4 |
| GGCT | SDC1 | 0.689981 | 6 | 0 | 6 |
| GGCT | MARVELD2 | 0.681115 | 5 | 0 | 4 |
| GGCT | CMTM4 | 0.671284 | 3 | 0 | 3 |
| GGCT | BNC1 | 0.67051 | 4 | 0 | 3 |
| GGCT | PIGW | 0.664811 | 4 | 0 | 3 |
| GGCT | SDHC | 0.659055 | 4 | 0 | 3 |
| GGCT | MAP2K4 | 0.656252 | 3 | 0 | 3 |
| GGCT | DMKN | 0.654112 | 5 | 0 | 5 |
| GGCT | SERBP1 | 0.649479 | 5 | 0 | 4 |
| GGCT | PLEK2 | 0.646506 | 4 | 0 | 4 |
| GGCT | MAL2 | 0.646409 | 6 | 0 | 6 |
| GGCT | CELSR1 | 0.642308 | 5 | 0 | 4 |
| GGCT | SQLE | 0.641692 | 7 | 0 | 6 |
| GGCT | CXADR | 0.640219 | 7 | 0 | 5 |
| GGCT | CDH1 | 0.631881 | 9 | 0 | 6 |
| GGCT | F11R | 0.631763 | 8 | 0 | 5 |
| GGCT | REEP4 | 0.631573 | 6 | 0 | 4 |
| GGCT | FAM83C | 0.63086 | 4 | 0 | 4 |
| GGCT | SORD | 0.629449 | 8 | 0 | 6 |
For details and further investigation, click here