| Full name: slingshot protein phosphatase 2 | Alias Symbol: KIAA1725 | ||
| Type: protein-coding gene | Cytoband: 17q11.2 | ||
| Entrez ID: 85464 | HGNC ID: HGNC:30580 | Ensembl Gene: ENSG00000141298 | OMIM ID: 606779 |
| Drug and gene relationship at DGIdb | |||
SSH2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of SSH2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SSH2 | 85464 | 226080_at | 0.5613 | 0.3093 | |
| GSE26886 | SSH2 | 85464 | 226080_at | 0.0226 | 0.9278 | |
| GSE45670 | SSH2 | 85464 | 226080_at | 0.1841 | 0.3387 | |
| GSE53622 | SSH2 | 85464 | 34769 | -0.1137 | 0.2981 | |
| GSE53624 | SSH2 | 85464 | 34769 | 0.0837 | 0.2757 | |
| GSE63941 | SSH2 | 85464 | 226080_at | -0.3778 | 0.5965 | |
| GSE77861 | SSH2 | 85464 | 226080_at | 0.1430 | 0.6267 | |
| GSE97050 | SSH2 | 85464 | A_33_P3367917 | 0.3487 | 0.2066 | |
| SRP007169 | SSH2 | 85464 | RNAseq | 1.1519 | 0.0352 | |
| SRP008496 | SSH2 | 85464 | RNAseq | 1.6361 | 0.0000 | |
| SRP064894 | SSH2 | 85464 | RNAseq | 0.5695 | 0.0080 | |
| SRP133303 | SSH2 | 85464 | RNAseq | 0.2510 | 0.0583 | |
| SRP159526 | SSH2 | 85464 | RNAseq | -0.0763 | 0.8409 | |
| SRP193095 | SSH2 | 85464 | RNAseq | 0.5299 | 0.0009 | |
| SRP219564 | SSH2 | 85464 | RNAseq | 0.4008 | 0.3781 | |
| TCGA | SSH2 | 85464 | RNAseq | -0.0655 | 0.3497 |
Upregulated datasets: 2; Downregulated datasets: 0.
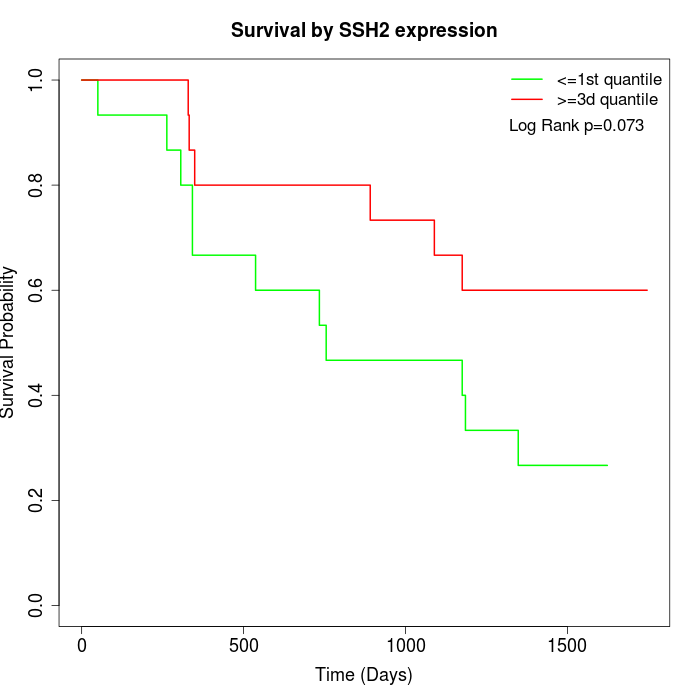
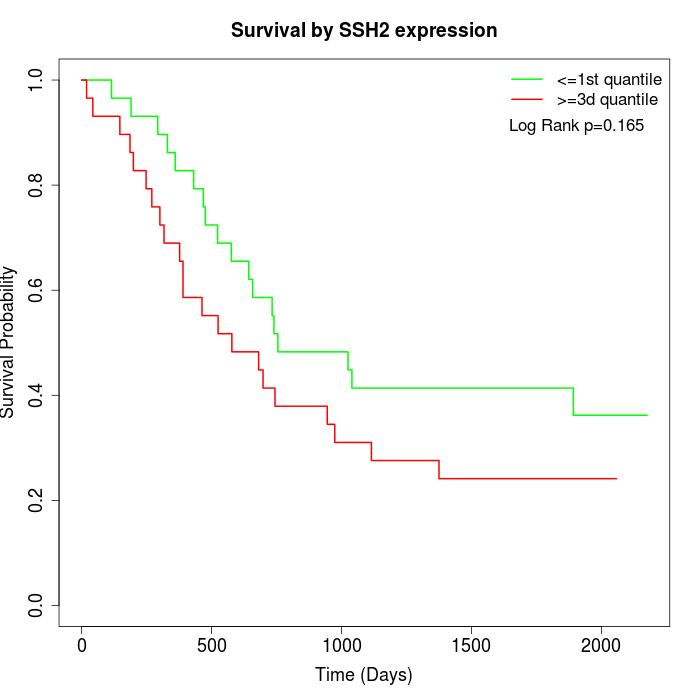
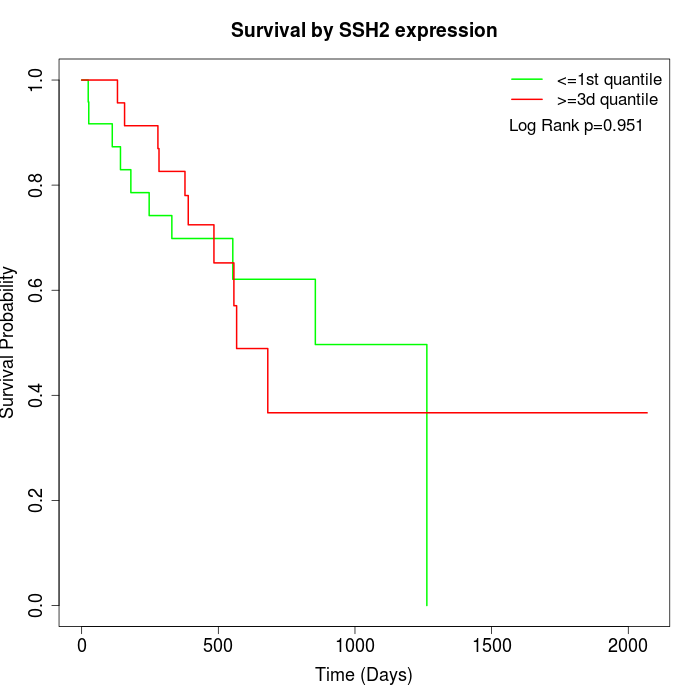
Survival by SSH2 expression:
Note: Click image to view full size file.
Copy number change of SSH2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SSH2 | 85464 | 7 | 2 | 21 | |
| GSE20123 | SSH2 | 85464 | 8 | 2 | 20 | |
| GSE43470 | SSH2 | 85464 | 0 | 1 | 42 | |
| GSE46452 | SSH2 | 85464 | 33 | 1 | 25 | |
| GSE47630 | SSH2 | 85464 | 7 | 1 | 32 | |
| GSE54993 | SSH2 | 85464 | 2 | 3 | 65 | |
| GSE54994 | SSH2 | 85464 | 7 | 5 | 41 | |
| GSE60625 | SSH2 | 85464 | 4 | 0 | 7 | |
| GSE74703 | SSH2 | 85464 | 0 | 1 | 35 | |
| GSE74704 | SSH2 | 85464 | 5 | 2 | 13 | |
| TCGA | SSH2 | 85464 | 22 | 9 | 65 |
Total number of gains: 95; Total number of losses: 27; Total Number of normals: 366.
Somatic mutations of SSH2:
Generating mutation plots.
Highly correlated genes for SSH2:
Showing top 20/152 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SSH2 | ACVRL1 | 0.769422 | 3 | 0 | 3 |
| SSH2 | EDEM1 | 0.757965 | 3 | 0 | 3 |
| SSH2 | PHTF1 | 0.74991 | 3 | 0 | 3 |
| SSH2 | MOGS | 0.738015 | 3 | 0 | 3 |
| SSH2 | SOX18 | 0.73715 | 3 | 0 | 3 |
| SSH2 | MSH6 | 0.733549 | 3 | 0 | 3 |
| SSH2 | MTHFD2 | 0.728091 | 3 | 0 | 3 |
| SSH2 | MMP3 | 0.720971 | 3 | 0 | 3 |
| SSH2 | CCT5 | 0.719219 | 3 | 0 | 3 |
| SSH2 | FAM167B | 0.717596 | 3 | 0 | 3 |
| SSH2 | USP39 | 0.715403 | 3 | 0 | 3 |
| SSH2 | ARHGAP9 | 0.711241 | 4 | 0 | 4 |
| SSH2 | AKR1B1 | 0.7077 | 3 | 0 | 3 |
| SSH2 | GLIPR1 | 0.703943 | 3 | 0 | 3 |
| SSH2 | FZR1 | 0.70184 | 3 | 0 | 3 |
| SSH2 | SPAST | 0.700096 | 3 | 0 | 3 |
| SSH2 | NAGLU | 0.700088 | 4 | 0 | 4 |
| SSH2 | POLR2A | 0.69648 | 3 | 0 | 3 |
| SSH2 | KIAA0319L | 0.695948 | 3 | 0 | 3 |
| SSH2 | TNFAIP8L2 | 0.69527 | 3 | 0 | 3 |
For details and further investigation, click here