| Full name: tachykinin precursor 1 | Alias Symbol: NPK | ||
| Type: protein-coding gene | Cytoband: 7q21.3 | ||
| Entrez ID: 6863 | HGNC ID: HGNC:11517 | Ensembl Gene: ENSG00000006128 | OMIM ID: 162320 |
| Drug and gene relationship at DGIdb | |||
Expression of TAC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TAC1 | 6863 | 206552_s_at | -0.3008 | 0.6328 | |
| GSE20347 | TAC1 | 6863 | 206552_s_at | 0.0592 | 0.4192 | |
| GSE23400 | TAC1 | 6863 | 206552_s_at | -0.0568 | 0.0179 | |
| GSE26886 | TAC1 | 6863 | 206552_s_at | 0.0650 | 0.4408 | |
| GSE29001 | TAC1 | 6863 | 206552_s_at | -0.0668 | 0.7255 | |
| GSE38129 | TAC1 | 6863 | 206552_s_at | -0.4726 | 0.0755 | |
| GSE45670 | TAC1 | 6863 | 206552_s_at | -0.1203 | 0.0760 | |
| GSE53622 | TAC1 | 6863 | 51225 | -0.0311 | 0.9077 | |
| GSE53624 | TAC1 | 6863 | 51225 | -0.0424 | 0.8013 | |
| GSE63941 | TAC1 | 6863 | 206552_s_at | -0.5411 | 0.0153 | |
| GSE77861 | TAC1 | 6863 | 206552_s_at | 0.0182 | 0.8441 | |
| TCGA | TAC1 | 6863 | RNAseq | -2.1858 | 0.0874 |
Upregulated datasets: 0; Downregulated datasets: 0.
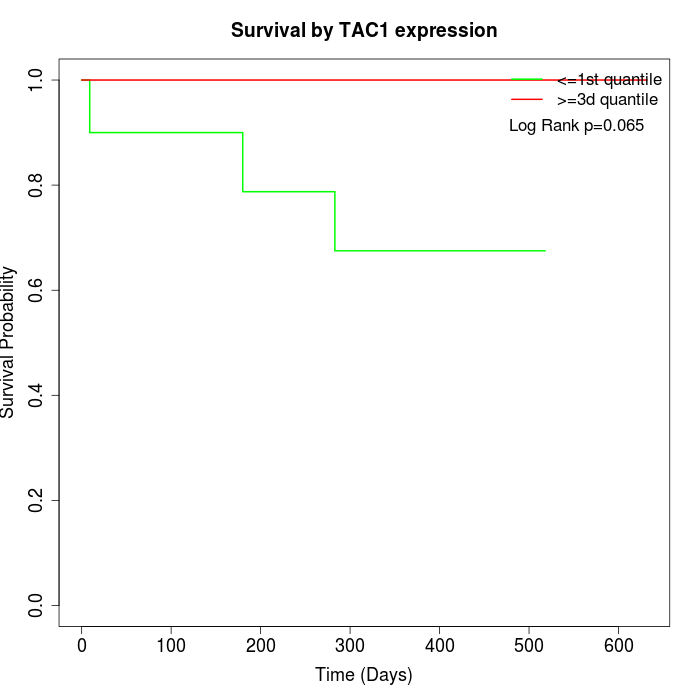
Survival by TAC1 expression:
Note: Click image to view full size file.
Copy number change of TAC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TAC1 | 6863 | 13 | 0 | 17 | |
| GSE20123 | TAC1 | 6863 | 13 | 0 | 17 | |
| GSE43470 | TAC1 | 6863 | 7 | 1 | 35 | |
| GSE46452 | TAC1 | 6863 | 12 | 0 | 47 | |
| GSE47630 | TAC1 | 6863 | 9 | 2 | 29 | |
| GSE54993 | TAC1 | 6863 | 1 | 8 | 61 | |
| GSE54994 | TAC1 | 6863 | 16 | 3 | 34 | |
| GSE60625 | TAC1 | 6863 | 0 | 5 | 6 | |
| GSE74703 | TAC1 | 6863 | 6 | 1 | 29 | |
| GSE74704 | TAC1 | 6863 | 9 | 0 | 11 | |
| TCGA | TAC1 | 6863 | 54 | 5 | 37 |
Total number of gains: 140; Total number of losses: 25; Total Number of normals: 323.
Somatic mutations of TAC1:
Generating mutation plots.
Highly correlated genes for TAC1:
Showing top 20/225 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TAC1 | FZD4 | 0.734821 | 3 | 0 | 3 |
| TAC1 | GDF10 | 0.731793 | 3 | 0 | 3 |
| TAC1 | MGARP | 0.685677 | 4 | 0 | 4 |
| TAC1 | PCDH9 | 0.68561 | 5 | 0 | 5 |
| TAC1 | ANGPTL7 | 0.683914 | 5 | 0 | 4 |
| TAC1 | ARID5A | 0.671087 | 4 | 0 | 3 |
| TAC1 | EDNRB | 0.656157 | 5 | 0 | 4 |
| TAC1 | NPR2 | 0.654964 | 5 | 0 | 3 |
| TAC1 | C20orf194 | 0.650938 | 3 | 0 | 3 |
| TAC1 | PCK1 | 0.644507 | 4 | 0 | 3 |
| TAC1 | THRA | 0.644267 | 3 | 0 | 3 |
| TAC1 | FENDRR | 0.644042 | 4 | 0 | 3 |
| TAC1 | CDC42EP2 | 0.63811 | 5 | 0 | 4 |
| TAC1 | CORO2B | 0.632795 | 6 | 0 | 4 |
| TAC1 | DGKG | 0.632266 | 4 | 0 | 3 |
| TAC1 | NR4A1 | 0.630553 | 4 | 0 | 3 |
| TAC1 | PKD2 | 0.627014 | 4 | 0 | 3 |
| TAC1 | GMPR | 0.626667 | 3 | 0 | 3 |
| TAC1 | FGF13-AS1 | 0.623792 | 3 | 0 | 3 |
| TAC1 | KLF2 | 0.620857 | 6 | 0 | 5 |
For details and further investigation, click here