| Full name: TNF receptor superfamily member 10d | Alias Symbol: DcR2|TRUNDD|TRAILR4|CD264 | ||
| Type: protein-coding gene | Cytoband: 8p21.3 | ||
| Entrez ID: 8793 | HGNC ID: HGNC:11907 | Ensembl Gene: ENSG00000173530 | OMIM ID: 603614 |
| Drug and gene relationship at DGIdb | |||
TNFRSF10D involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04210 | Apoptosis | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa05162 | Measles |
Expression of TNFRSF10D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNFRSF10D | 8793 | 227345_at | 0.5041 | 0.4970 | |
| GSE20347 | TNFRSF10D | 8793 | 210654_at | 0.0666 | 0.2167 | |
| GSE23400 | TNFRSF10D | 8793 | 210654_at | -0.0080 | 0.7795 | |
| GSE26886 | TNFRSF10D | 8793 | 227345_at | 0.7354 | 0.0977 | |
| GSE29001 | TNFRSF10D | 8793 | 210654_at | 0.0295 | 0.8484 | |
| GSE38129 | TNFRSF10D | 8793 | 210654_at | 0.0295 | 0.6353 | |
| GSE45670 | TNFRSF10D | 8793 | 227345_at | 0.5074 | 0.1927 | |
| GSE53622 | TNFRSF10D | 8793 | 12276 | 0.4102 | 0.0493 | |
| GSE53624 | TNFRSF10D | 8793 | 12276 | 0.5936 | 0.0001 | |
| GSE63941 | TNFRSF10D | 8793 | 227345_at | -2.9020 | 0.0450 | |
| GSE77861 | TNFRSF10D | 8793 | 227345_at | 0.8779 | 0.2690 | |
| GSE97050 | TNFRSF10D | 8793 | A_33_P3326588 | 0.2353 | 0.6631 | |
| SRP007169 | TNFRSF10D | 8793 | RNAseq | 0.7505 | 0.2511 | |
| SRP064894 | TNFRSF10D | 8793 | RNAseq | 0.4667 | 0.1152 | |
| SRP133303 | TNFRSF10D | 8793 | RNAseq | 0.6217 | 0.0288 | |
| SRP159526 | TNFRSF10D | 8793 | RNAseq | 0.4675 | 0.3948 | |
| SRP193095 | TNFRSF10D | 8793 | RNAseq | 1.2822 | 0.0000 | |
| SRP219564 | TNFRSF10D | 8793 | RNAseq | -0.9073 | 0.1850 | |
| TCGA | TNFRSF10D | 8793 | RNAseq | -0.4119 | 0.0318 |
Upregulated datasets: 1; Downregulated datasets: 1.
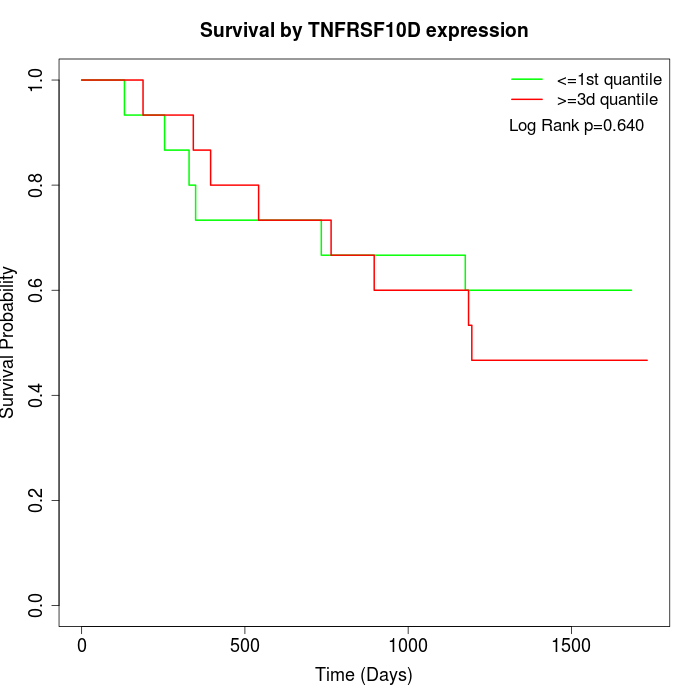
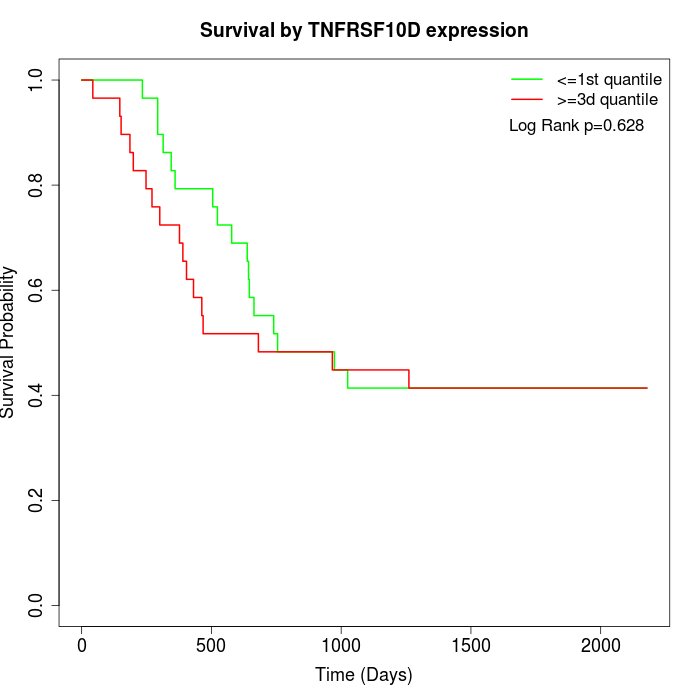
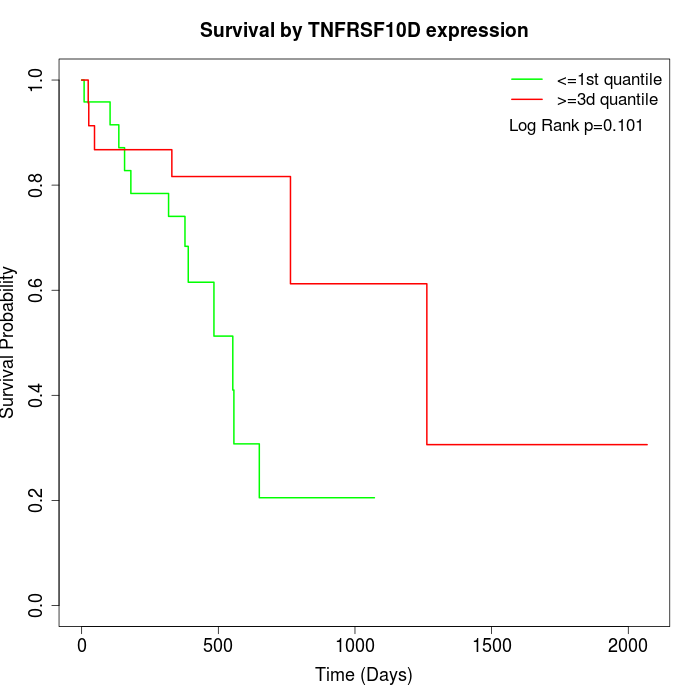
Survival by TNFRSF10D expression:
Note: Click image to view full size file.
Copy number change of TNFRSF10D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNFRSF10D | 8793 | 4 | 12 | 14 | |
| GSE20123 | TNFRSF10D | 8793 | 4 | 12 | 14 | |
| GSE43470 | TNFRSF10D | 8793 | 4 | 9 | 30 | |
| GSE46452 | TNFRSF10D | 8793 | 13 | 13 | 33 | |
| GSE47630 | TNFRSF10D | 8793 | 10 | 8 | 22 | |
| GSE54993 | TNFRSF10D | 8793 | 2 | 14 | 54 | |
| GSE54994 | TNFRSF10D | 8793 | 8 | 18 | 27 | |
| GSE60625 | TNFRSF10D | 8793 | 3 | 0 | 8 | |
| GSE74703 | TNFRSF10D | 8793 | 4 | 7 | 25 | |
| GSE74704 | TNFRSF10D | 8793 | 3 | 8 | 9 | |
| TCGA | TNFRSF10D | 8793 | 14 | 43 | 39 |
Total number of gains: 69; Total number of losses: 144; Total Number of normals: 275.
Somatic mutations of TNFRSF10D:
Generating mutation plots.
Highly correlated genes for TNFRSF10D:
Showing top 20/510 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNFRSF10D | LRRC34 | 0.766922 | 3 | 0 | 3 |
| TNFRSF10D | LCAT | 0.762364 | 3 | 0 | 3 |
| TNFRSF10D | HMCN1 | 0.762088 | 4 | 0 | 4 |
| TNFRSF10D | DCHS1 | 0.742333 | 3 | 0 | 3 |
| TNFRSF10D | TNFRSF19 | 0.740761 | 3 | 0 | 3 |
| TNFRSF10D | MRGPRF | 0.738954 | 3 | 0 | 3 |
| TNFRSF10D | FGF1 | 0.734492 | 3 | 0 | 3 |
| TNFRSF10D | MAN1A1 | 0.732446 | 4 | 0 | 3 |
| TNFRSF10D | SPRY4-IT1 | 0.730027 | 4 | 0 | 4 |
| TNFRSF10D | STK17A | 0.72828 | 4 | 0 | 3 |
| TNFRSF10D | GNL3L | 0.726411 | 3 | 0 | 3 |
| TNFRSF10D | FZD1 | 0.725761 | 3 | 0 | 3 |
| TNFRSF10D | PCSK1 | 0.725646 | 3 | 0 | 3 |
| TNFRSF10D | KANK4 | 0.71975 | 4 | 0 | 3 |
| TNFRSF10D | MAGEH1 | 0.709517 | 3 | 0 | 3 |
| TNFRSF10D | PSMC1 | 0.708141 | 4 | 0 | 3 |
| TNFRSF10D | PPP1R21 | 0.705496 | 3 | 0 | 3 |
| TNFRSF10D | STX16 | 0.703519 | 3 | 0 | 3 |
| TNFRSF10D | SPATA9 | 0.69955 | 3 | 0 | 3 |
| TNFRSF10D | RPS27L | 0.698536 | 3 | 0 | 3 |
For details and further investigation, click here