| Full name: TNF receptor superfamily member 25 | Alias Symbol: DR3|TRAMP|WSL-1|LARD|WSL-LR|DDR3|TR3|APO-3 | ||
| Type: protein-coding gene | Cytoband: 1p36.31 | ||
| Entrez ID: 8718 | HGNC ID: HGNC:11910 | Ensembl Gene: ENSG00000215788 | OMIM ID: 603366 |
| Drug and gene relationship at DGIdb | |||
Expression of TNFRSF25:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNFRSF25 | 8718 | 211282_x_at | 0.9141 | 0.2514 | |
| GSE20347 | TNFRSF25 | 8718 | 211282_x_at | 0.1809 | 0.3376 | |
| GSE23400 | TNFRSF25 | 8718 | 216042_at | 0.0493 | 0.2653 | |
| GSE26886 | TNFRSF25 | 8718 | 216042_at | 0.0063 | 0.9658 | |
| GSE29001 | TNFRSF25 | 8718 | 219423_x_at | 0.2401 | 0.5396 | |
| GSE38129 | TNFRSF25 | 8718 | 211282_x_at | 0.3255 | 0.0285 | |
| GSE45670 | TNFRSF25 | 8718 | 219423_x_at | 0.5933 | 0.0084 | |
| GSE53622 | TNFRSF25 | 8718 | 13449 | 0.2807 | 0.0212 | |
| GSE53624 | TNFRSF25 | 8718 | 71558 | 0.3824 | 0.0003 | |
| GSE63941 | TNFRSF25 | 8718 | 211282_x_at | 0.3536 | 0.0216 | |
| GSE77861 | TNFRSF25 | 8718 | 216042_at | 0.0319 | 0.7949 | |
| GSE97050 | TNFRSF25 | 8718 | A_23_P126844 | 0.1587 | 0.6565 | |
| SRP064894 | TNFRSF25 | 8718 | RNAseq | 0.6458 | 0.0052 | |
| SRP133303 | TNFRSF25 | 8718 | RNAseq | -0.0869 | 0.5697 | |
| SRP159526 | TNFRSF25 | 8718 | RNAseq | -0.1038 | 0.6944 | |
| SRP193095 | TNFRSF25 | 8718 | RNAseq | 0.4431 | 0.0315 | |
| SRP219564 | TNFRSF25 | 8718 | RNAseq | -0.1670 | 0.7249 | |
| TCGA | TNFRSF25 | 8718 | RNAseq | 0.6963 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
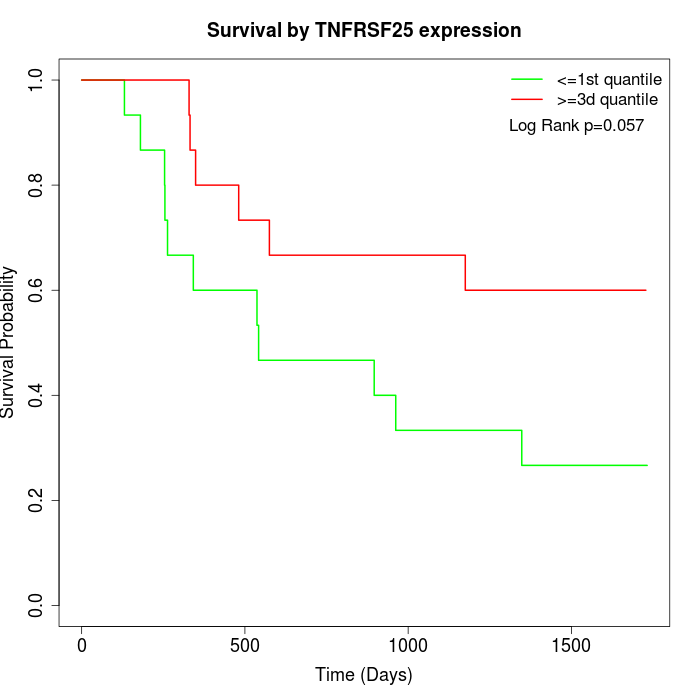
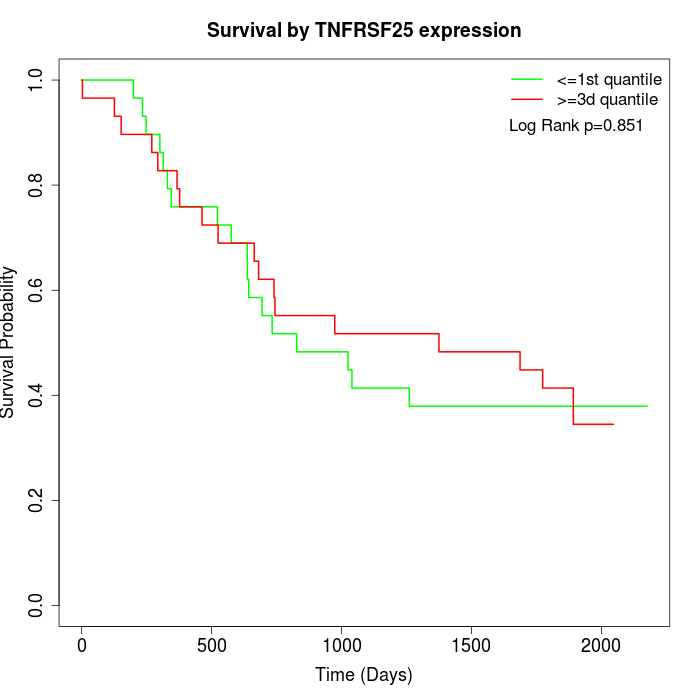
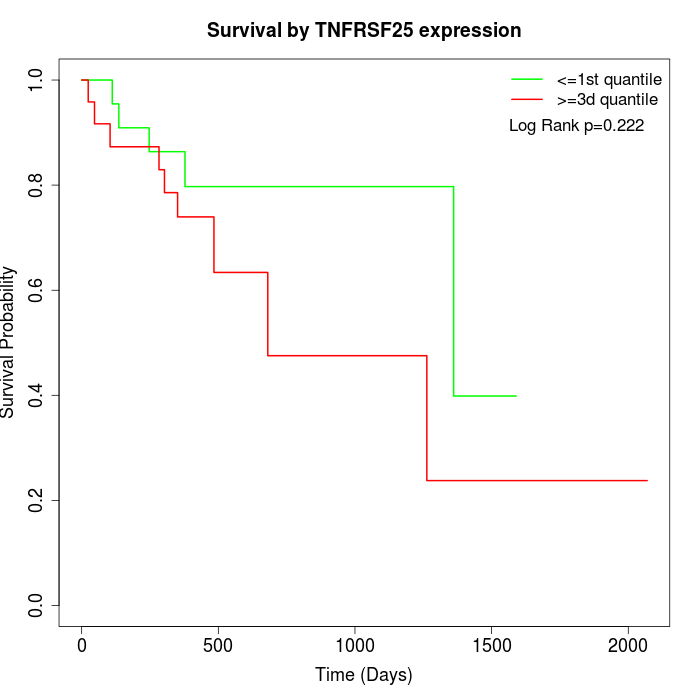
Survival by TNFRSF25 expression:
Note: Click image to view full size file.
Copy number change of TNFRSF25:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNFRSF25 | 8718 | 3 | 6 | 21 | |
| GSE20123 | TNFRSF25 | 8718 | 3 | 5 | 22 | |
| GSE43470 | TNFRSF25 | 8718 | 6 | 5 | 32 | |
| GSE46452 | TNFRSF25 | 8718 | 7 | 1 | 51 | |
| GSE47630 | TNFRSF25 | 8718 | 8 | 4 | 28 | |
| GSE54993 | TNFRSF25 | 8718 | 3 | 2 | 65 | |
| GSE54994 | TNFRSF25 | 8718 | 14 | 3 | 36 | |
| GSE60625 | TNFRSF25 | 8718 | 0 | 0 | 11 | |
| GSE74703 | TNFRSF25 | 8718 | 5 | 3 | 28 | |
| GSE74704 | TNFRSF25 | 8718 | 3 | 0 | 17 | |
| TCGA | TNFRSF25 | 8718 | 12 | 22 | 62 |
Total number of gains: 64; Total number of losses: 51; Total Number of normals: 373.
Somatic mutations of TNFRSF25:
Generating mutation plots.
Highly correlated genes for TNFRSF25:
Showing top 20/210 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNFRSF25 | ZAN | 0.803139 | 3 | 0 | 3 |
| TNFRSF25 | GZMH | 0.70999 | 3 | 0 | 3 |
| TNFRSF25 | HELZ2 | 0.709603 | 3 | 0 | 3 |
| TNFRSF25 | ZBTB17 | 0.703682 | 6 | 0 | 6 |
| TNFRSF25 | FGFR4 | 0.70263 | 3 | 0 | 3 |
| TNFRSF25 | TUBGCP6 | 0.698632 | 3 | 0 | 3 |
| TNFRSF25 | POLR3A | 0.686436 | 3 | 0 | 3 |
| TNFRSF25 | MEGF10 | 0.681462 | 3 | 0 | 3 |
| TNFRSF25 | INF2 | 0.670731 | 5 | 0 | 5 |
| TNFRSF25 | ZNF341 | 0.670298 | 4 | 0 | 3 |
| TNFRSF25 | TRIM41 | 0.669589 | 4 | 0 | 4 |
| TNFRSF25 | PARS2 | 0.658259 | 3 | 0 | 3 |
| TNFRSF25 | ZNF526 | 0.657814 | 3 | 0 | 3 |
| TNFRSF25 | CCDC81 | 0.65757 | 4 | 0 | 3 |
| TNFRSF25 | NLRC5 | 0.656317 | 5 | 0 | 5 |
| TNFRSF25 | HAUS5 | 0.648277 | 8 | 0 | 7 |
| TNFRSF25 | KIFC2 | 0.646069 | 4 | 0 | 3 |
| TNFRSF25 | CDK5R2 | 0.646009 | 4 | 0 | 3 |
| TNFRSF25 | TSR1 | 0.640778 | 6 | 0 | 5 |
| TNFRSF25 | FABP6 | 0.633135 | 5 | 0 | 4 |
For details and further investigation, click here