| Full name: TRAF2 and NCK interacting kinase | Alias Symbol: KIAA0551 | ||
| Type: protein-coding gene | Cytoband: 3q26.2-q26.31 | ||
| Entrez ID: 23043 | HGNC ID: HGNC:30765 | Ensembl Gene: ENSG00000154310 | OMIM ID: 610005 |
| Drug and gene relationship at DGIdb | |||
Expression of TNIK:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNIK | 23043 | 211828_s_at | -0.8532 | 0.3133 | |
| GSE20347 | TNIK | 23043 | 213107_at | -0.0595 | 0.8124 | |
| GSE23400 | TNIK | 23043 | 213107_at | -0.1424 | 0.0172 | |
| GSE26886 | TNIK | 23043 | 213107_at | -0.4598 | 0.1368 | |
| GSE29001 | TNIK | 23043 | 213107_at | -0.3019 | 0.3692 | |
| GSE38129 | TNIK | 23043 | 213107_at | -0.2280 | 0.2863 | |
| GSE45670 | TNIK | 23043 | 213107_at | -0.7724 | 0.0281 | |
| GSE53622 | TNIK | 23043 | 83556 | -0.5800 | 0.0000 | |
| GSE53624 | TNIK | 23043 | 83556 | -0.5234 | 0.0000 | |
| GSE63941 | TNIK | 23043 | 213107_at | -1.5032 | 0.1319 | |
| GSE77861 | TNIK | 23043 | 213107_at | 0.0197 | 0.9427 | |
| GSE97050 | TNIK | 23043 | A_33_P3213596 | -0.7260 | 0.2415 | |
| SRP007169 | TNIK | 23043 | RNAseq | 2.1939 | 0.0057 | |
| SRP064894 | TNIK | 23043 | RNAseq | -0.1121 | 0.5562 | |
| SRP133303 | TNIK | 23043 | RNAseq | -0.1865 | 0.4951 | |
| SRP159526 | TNIK | 23043 | RNAseq | -0.3267 | 0.3668 | |
| SRP193095 | TNIK | 23043 | RNAseq | -0.2002 | 0.5428 | |
| SRP219564 | TNIK | 23043 | RNAseq | -0.3496 | 0.6000 | |
| TCGA | TNIK | 23043 | RNAseq | -0.6416 | 0.0019 |
Upregulated datasets: 1; Downregulated datasets: 0.
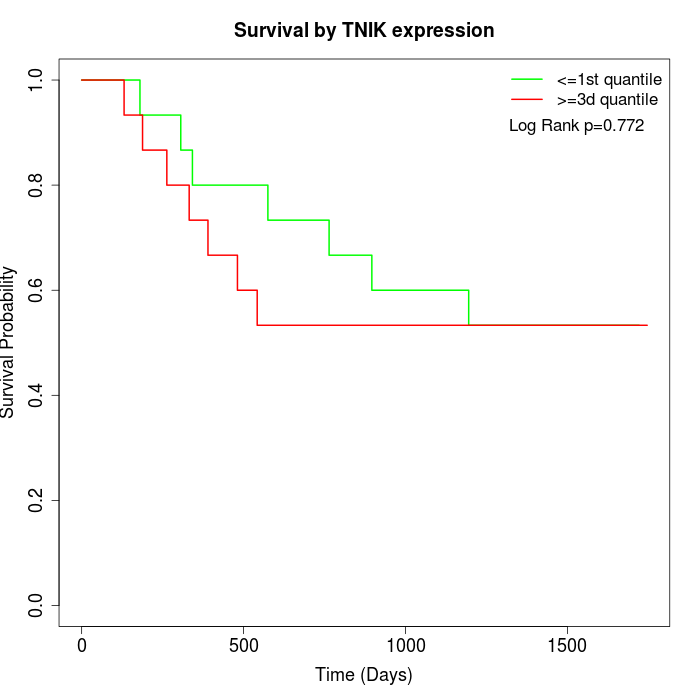
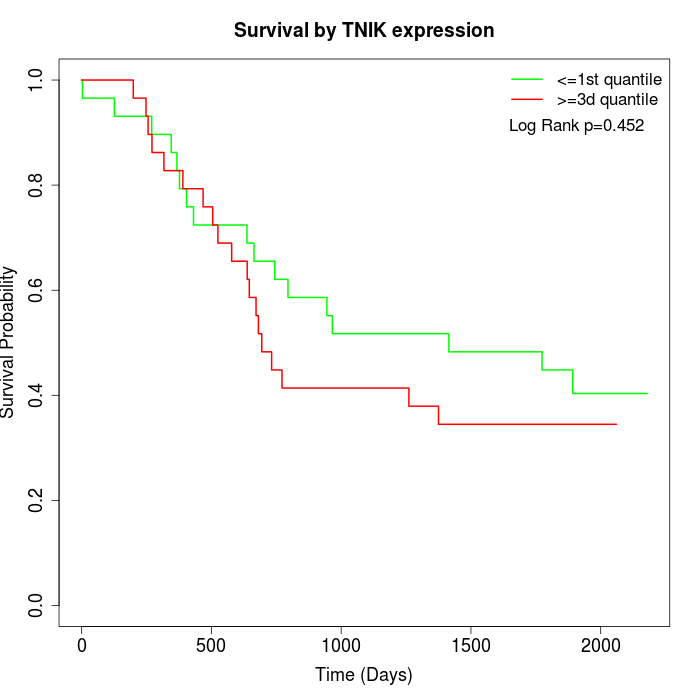
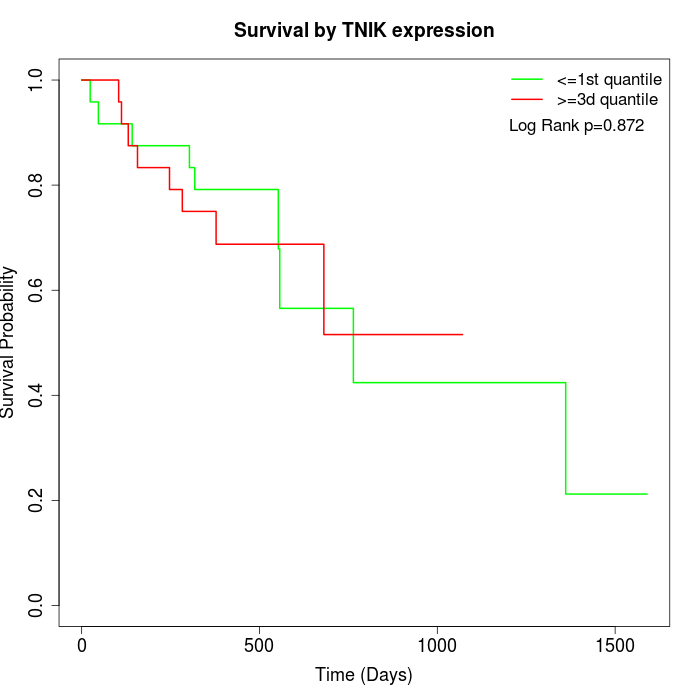
Survival by TNIK expression:
Note: Click image to view full size file.
Copy number change of TNIK:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNIK | 23043 | 24 | 0 | 6 | |
| GSE20123 | TNIK | 23043 | 24 | 0 | 6 | |
| GSE43470 | TNIK | 23043 | 24 | 0 | 19 | |
| GSE46452 | TNIK | 23043 | 19 | 1 | 39 | |
| GSE47630 | TNIK | 23043 | 27 | 2 | 11 | |
| GSE54993 | TNIK | 23043 | 1 | 15 | 54 | |
| GSE54994 | TNIK | 23043 | 41 | 0 | 12 | |
| GSE60625 | TNIK | 23043 | 0 | 6 | 5 | |
| GSE74703 | TNIK | 23043 | 20 | 0 | 16 | |
| GSE74704 | TNIK | 23043 | 16 | 0 | 4 | |
| TCGA | TNIK | 23043 | 80 | 0 | 16 |
Total number of gains: 276; Total number of losses: 24; Total Number of normals: 188.
Somatic mutations of TNIK:
Generating mutation plots.
Highly correlated genes for TNIK:
Showing top 20/382 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNIK | COL4A6 | 0.734886 | 3 | 0 | 3 |
| TNIK | CCDC136 | 0.727082 | 3 | 0 | 3 |
| TNIK | SLC2A12 | 0.724147 | 3 | 0 | 3 |
| TNIK | ARHGEF25 | 0.719622 | 4 | 0 | 4 |
| TNIK | SLC35F1 | 0.715723 | 3 | 0 | 3 |
| TNIK | KATNAL1 | 0.715242 | 4 | 0 | 4 |
| TNIK | PRR26 | 0.696005 | 3 | 0 | 3 |
| TNIK | IQUB | 0.692671 | 3 | 0 | 3 |
| TNIK | GIPC3 | 0.685099 | 3 | 0 | 3 |
| TNIK | STK33 | 0.668373 | 6 | 0 | 5 |
| TNIK | THRA | 0.657687 | 4 | 0 | 3 |
| TNIK | LRRTM1 | 0.654844 | 4 | 0 | 3 |
| TNIK | NRXN3 | 0.654832 | 6 | 0 | 5 |
| TNIK | CDHR3 | 0.653035 | 4 | 0 | 3 |
| TNIK | SMAD9 | 0.647132 | 6 | 0 | 4 |
| TNIK | JAZF1 | 0.639039 | 5 | 0 | 3 |
| TNIK | PTCHD1 | 0.636801 | 6 | 0 | 4 |
| TNIK | PTPDC1 | 0.635975 | 3 | 0 | 3 |
| TNIK | GKAP1 | 0.635087 | 4 | 0 | 3 |
| TNIK | NEXN | 0.633553 | 5 | 0 | 3 |
For details and further investigation, click here