| Full name: tensin 1 | Alias Symbol: DKFZp586K0617|PPP1R155 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 7145 | HGNC ID: HGNC:11973 | Ensembl Gene: ENSG00000079308 | OMIM ID: 600076 |
| Drug and gene relationship at DGIdb | |||
Expression of TNS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNS1 | 7145 | 221748_s_at | -1.6151 | 0.1217 | |
| GSE20347 | TNS1 | 7145 | 221748_s_at | -0.7631 | 0.0333 | |
| GSE23400 | TNS1 | 7145 | 221748_s_at | -0.8107 | 0.0001 | |
| GSE26886 | TNS1 | 7145 | 221748_s_at | 0.6358 | 0.1626 | |
| GSE29001 | TNS1 | 7145 | 221748_s_at | -0.8805 | 0.0445 | |
| GSE38129 | TNS1 | 7145 | 221748_s_at | -1.1399 | 0.0236 | |
| GSE45670 | TNS1 | 7145 | 221748_s_at | -2.5428 | 0.0000 | |
| GSE53622 | TNS1 | 7145 | 71531 | -1.8802 | 0.0000 | |
| GSE53624 | TNS1 | 7145 | 71531 | -1.6457 | 0.0000 | |
| GSE63941 | TNS1 | 7145 | 221246_x_at | -0.7541 | 0.1572 | |
| GSE77861 | TNS1 | 7145 | 221747_at | -0.4008 | 0.0409 | |
| GSE97050 | TNS1 | 7145 | A_33_P3209491 | -0.9812 | 0.1525 | |
| SRP007169 | TNS1 | 7145 | RNAseq | 1.2254 | 0.0005 | |
| SRP008496 | TNS1 | 7145 | RNAseq | 1.1575 | 0.0001 | |
| SRP064894 | TNS1 | 7145 | RNAseq | -0.5751 | 0.1255 | |
| SRP133303 | TNS1 | 7145 | RNAseq | -0.9155 | 0.1072 | |
| SRP159526 | TNS1 | 7145 | RNAseq | -1.1646 | 0.0308 | |
| SRP193095 | TNS1 | 7145 | RNAseq | -0.0079 | 0.9649 | |
| SRP219564 | TNS1 | 7145 | RNAseq | -0.5950 | 0.5507 | |
| TCGA | TNS1 | 7145 | RNAseq | -0.5280 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 5.
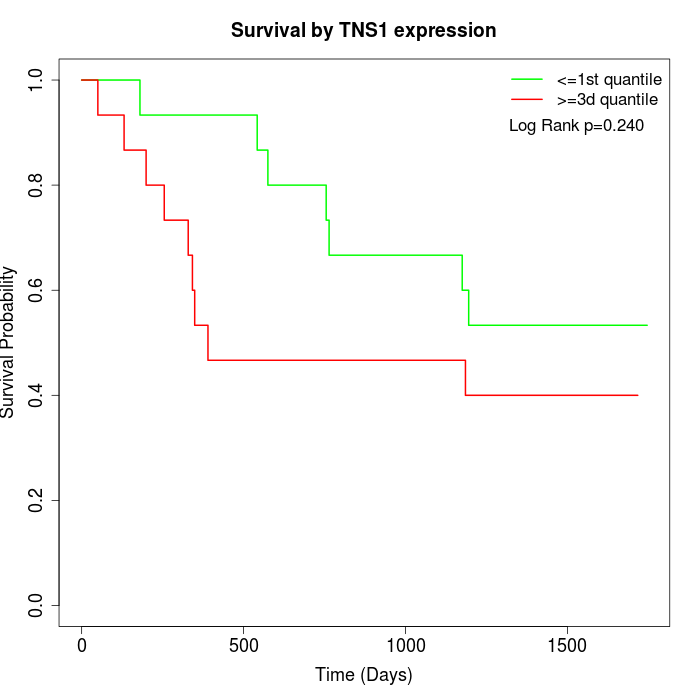
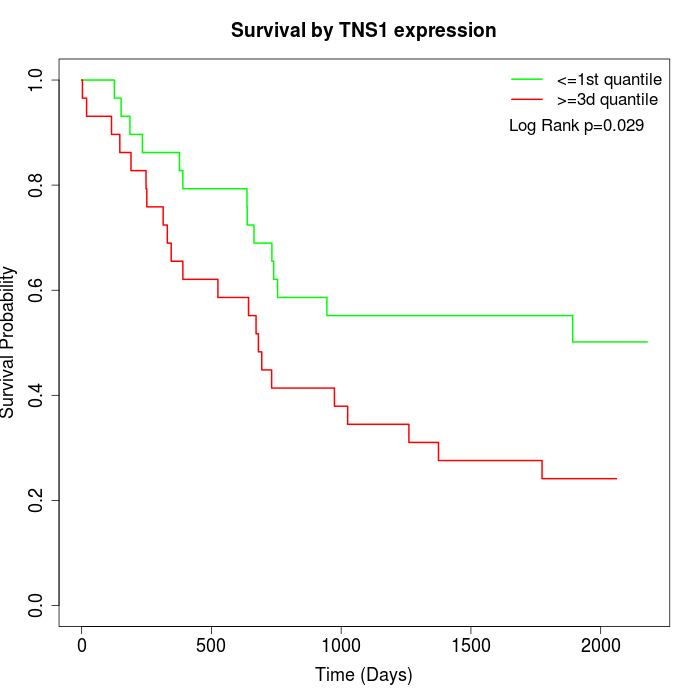
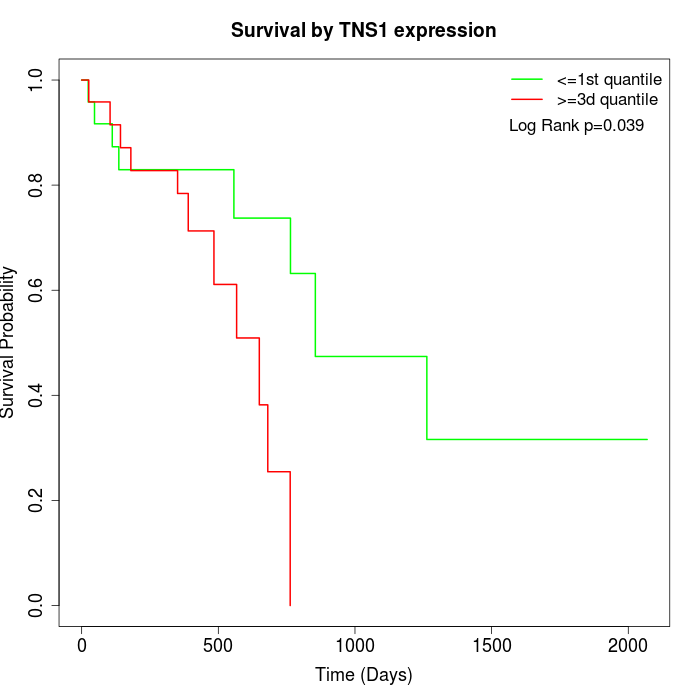
Survival by TNS1 expression:
Note: Click image to view full size file.
Copy number change of TNS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNS1 | 7145 | 1 | 13 | 16 | |
| GSE20123 | TNS1 | 7145 | 1 | 13 | 16 | |
| GSE43470 | TNS1 | 7145 | 1 | 7 | 35 | |
| GSE46452 | TNS1 | 7145 | 0 | 5 | 54 | |
| GSE47630 | TNS1 | 7145 | 4 | 5 | 31 | |
| GSE54993 | TNS1 | 7145 | 0 | 3 | 67 | |
| GSE54994 | TNS1 | 7145 | 8 | 10 | 35 | |
| GSE60625 | TNS1 | 7145 | 0 | 3 | 8 | |
| GSE74703 | TNS1 | 7145 | 1 | 5 | 30 | |
| GSE74704 | TNS1 | 7145 | 1 | 7 | 12 | |
| TCGA | TNS1 | 7145 | 11 | 27 | 58 |
Total number of gains: 28; Total number of losses: 98; Total Number of normals: 362.
Somatic mutations of TNS1:
Generating mutation plots.
Highly correlated genes for TNS1:
Showing top 20/1362 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNS1 | CYP21A2 | 0.87035 | 3 | 0 | 3 |
| TNS1 | XKR4 | 0.849695 | 5 | 0 | 5 |
| TNS1 | CNN1 | 0.841179 | 9 | 0 | 9 |
| TNS1 | C3orf70 | 0.834899 | 6 | 0 | 6 |
| TNS1 | C1QTNF7 | 0.834163 | 5 | 0 | 5 |
| TNS1 | CLEC3B | 0.828583 | 3 | 0 | 3 |
| TNS1 | ATP1A2 | 0.828449 | 9 | 0 | 9 |
| TNS1 | CBX7 | 0.826974 | 9 | 0 | 9 |
| TNS1 | AQP1 | 0.825946 | 9 | 0 | 9 |
| TNS1 | PPP1R14A | 0.825823 | 6 | 0 | 6 |
| TNS1 | CHRDL1 | 0.823089 | 9 | 0 | 8 |
| TNS1 | SMOC2 | 0.819632 | 6 | 0 | 6 |
| TNS1 | LINC01279 | 0.811171 | 4 | 0 | 4 |
| TNS1 | AOC3 | 0.805945 | 11 | 0 | 10 |
| TNS1 | MAMDC2 | 0.805808 | 6 | 0 | 5 |
| TNS1 | MRVI1 | 0.797014 | 8 | 0 | 8 |
| TNS1 | PTCHD1 | 0.794814 | 5 | 0 | 5 |
| TNS1 | ACTG2 | 0.791343 | 11 | 0 | 10 |
| TNS1 | MYOCD | 0.790087 | 8 | 0 | 7 |
| TNS1 | PLN | 0.789603 | 11 | 0 | 9 |
For details and further investigation, click here