| Full name: triggering receptor expressed on myeloid cells like 1 | Alias Symbol: TLT1|dJ238O23.3 | ||
| Type: protein-coding gene | Cytoband: 6p21.1 | ||
| Entrez ID: 340205 | HGNC ID: HGNC:20434 | Ensembl Gene: ENSG00000161911 | OMIM ID: 609714 |
| Drug and gene relationship at DGIdb | |||
Expression of TREML1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TREML1 | 340205 | 1555659_a_at | 0.0633 | 0.8779 | |
| GSE26886 | TREML1 | 340205 | 1555659_a_at | -0.3052 | 0.0448 | |
| GSE45670 | TREML1 | 340205 | 1555659_a_at | 0.2186 | 0.0733 | |
| GSE53622 | TREML1 | 340205 | 58484 | 0.7924 | 0.0000 | |
| GSE53624 | TREML1 | 340205 | 58484 | 0.5295 | 0.0000 | |
| GSE63941 | TREML1 | 340205 | 1555659_a_at | 0.1488 | 0.4475 | |
| GSE77861 | TREML1 | 340205 | 1555659_a_at | 0.0113 | 0.9394 | |
| GSE97050 | TREML1 | 340205 | A_33_P3381777 | 0.1606 | 0.6121 | |
| SRP219564 | TREML1 | 340205 | RNAseq | 3.2879 | 0.0000 | |
| TCGA | TREML1 | 340205 | RNAseq | 1.3386 | 0.0568 |
Upregulated datasets: 1; Downregulated datasets: 0.
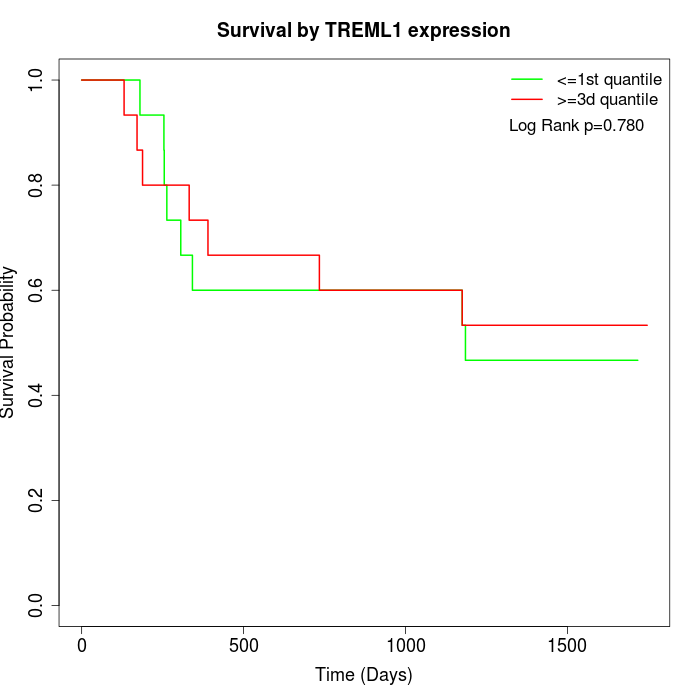
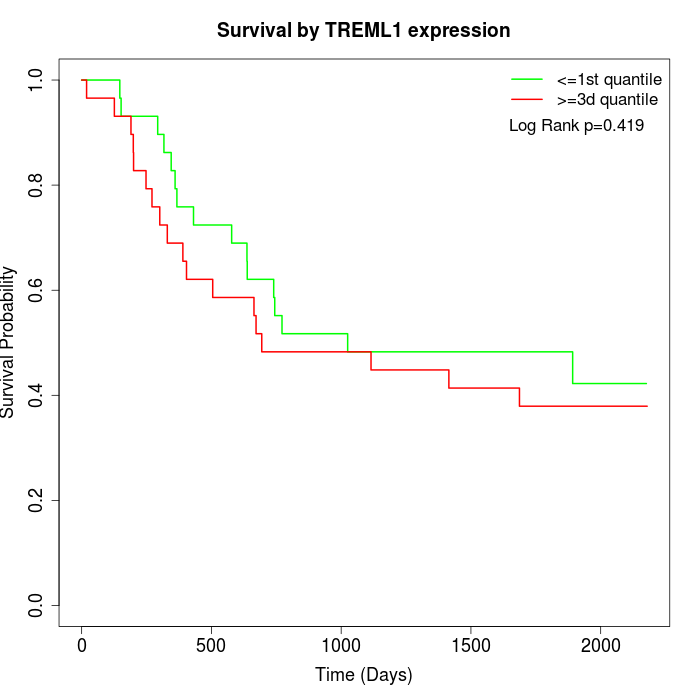
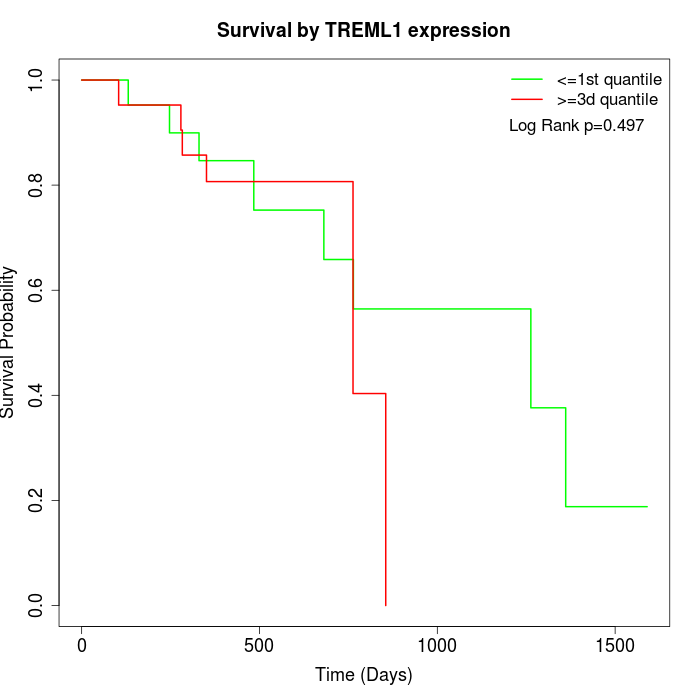
Survival by TREML1 expression:
Note: Click image to view full size file.
Copy number change of TREML1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TREML1 | 340205 | 3 | 3 | 24 | |
| GSE20123 | TREML1 | 340205 | 3 | 3 | 24 | |
| GSE43470 | TREML1 | 340205 | 6 | 0 | 37 | |
| GSE46452 | TREML1 | 340205 | 2 | 9 | 48 | |
| GSE47630 | TREML1 | 340205 | 8 | 4 | 28 | |
| GSE54993 | TREML1 | 340205 | 3 | 2 | 65 | |
| GSE54994 | TREML1 | 340205 | 11 | 4 | 38 | |
| GSE60625 | TREML1 | 340205 | 0 | 1 | 10 | |
| GSE74703 | TREML1 | 340205 | 6 | 0 | 30 | |
| GSE74704 | TREML1 | 340205 | 1 | 3 | 16 | |
| TCGA | TREML1 | 340205 | 19 | 13 | 64 |
Total number of gains: 62; Total number of losses: 42; Total Number of normals: 384.
Somatic mutations of TREML1:
Generating mutation plots.
Highly correlated genes for TREML1:
Showing top 20/69 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TREML1 | UNC5C | 0.694176 | 3 | 0 | 3 |
| TREML1 | PHACTR1 | 0.690826 | 3 | 0 | 3 |
| TREML1 | GNAT1 | 0.680534 | 3 | 0 | 3 |
| TREML1 | UAP1L1 | 0.673422 | 3 | 0 | 3 |
| TREML1 | ATG7 | 0.659292 | 3 | 0 | 3 |
| TREML1 | ZNF771 | 0.632381 | 3 | 0 | 3 |
| TREML1 | ENTPD2 | 0.632107 | 3 | 0 | 3 |
| TREML1 | AKT1S1 | 0.630113 | 4 | 0 | 3 |
| TREML1 | SCAMP2 | 0.619734 | 3 | 0 | 3 |
| TREML1 | UCMA | 0.616292 | 3 | 0 | 3 |
| TREML1 | RAB4B | 0.612713 | 4 | 0 | 3 |
| TREML1 | PDE8A | 0.608362 | 3 | 0 | 3 |
| TREML1 | SLC18A1 | 0.604644 | 3 | 0 | 3 |
| TREML1 | FN3K | 0.604196 | 4 | 0 | 4 |
| TREML1 | BTBD2 | 0.597979 | 4 | 0 | 3 |
| TREML1 | TUBG1 | 0.59672 | 3 | 0 | 3 |
| TREML1 | SERPINB11 | 0.596219 | 3 | 0 | 3 |
| TREML1 | TIRAP | 0.592141 | 5 | 0 | 5 |
| TREML1 | UBA1 | 0.5918 | 4 | 0 | 3 |
| TREML1 | TNFRSF10C | 0.591712 | 3 | 0 | 3 |
For details and further investigation, click here