| Full name: TIR domain containing adaptor protein | Alias Symbol: Mal|wyatt | ||
| Type: protein-coding gene | Cytoband: 11q24.2 | ||
| Entrez ID: 114609 | HGNC ID: HGNC:17192 | Ensembl Gene: ENSG00000150455 | OMIM ID: 606252 |
| Drug and gene relationship at DGIdb | |||
TIRAP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04620 | Toll-like receptor signaling pathway | |
| hsa05133 | Pertussis | |
| hsa05152 | Tuberculosis | |
| hsa05161 | Hepatitis B |
Expression of TIRAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TIRAP | 114609 | 1554091_a_at | -0.3519 | 0.1778 | |
| GSE26886 | TIRAP | 114609 | 1554091_a_at | -0.2491 | 0.1037 | |
| GSE45670 | TIRAP | 114609 | 1554091_a_at | -0.1021 | 0.2965 | |
| GSE53622 | TIRAP | 114609 | 41264 | -0.2678 | 0.0013 | |
| GSE53624 | TIRAP | 114609 | 41264 | -0.3131 | 0.0000 | |
| GSE63941 | TIRAP | 114609 | 1554091_a_at | 0.2299 | 0.4332 | |
| GSE77861 | TIRAP | 114609 | 1554091_a_at | -0.2259 | 0.1076 | |
| GSE97050 | TIRAP | 114609 | A_23_P397856 | -0.2570 | 0.2684 | |
| SRP007169 | TIRAP | 114609 | RNAseq | -1.6150 | 0.0001 | |
| SRP008496 | TIRAP | 114609 | RNAseq | -0.9668 | 0.0072 | |
| SRP064894 | TIRAP | 114609 | RNAseq | -0.3301 | 0.0839 | |
| SRP133303 | TIRAP | 114609 | RNAseq | -0.4089 | 0.0048 | |
| SRP159526 | TIRAP | 114609 | RNAseq | -0.1713 | 0.6281 | |
| SRP193095 | TIRAP | 114609 | RNAseq | -0.3564 | 0.0042 | |
| SRP219564 | TIRAP | 114609 | RNAseq | -0.3390 | 0.2762 | |
| TCGA | TIRAP | 114609 | RNAseq | -0.3181 | 0.0008 |
Upregulated datasets: 0; Downregulated datasets: 1.
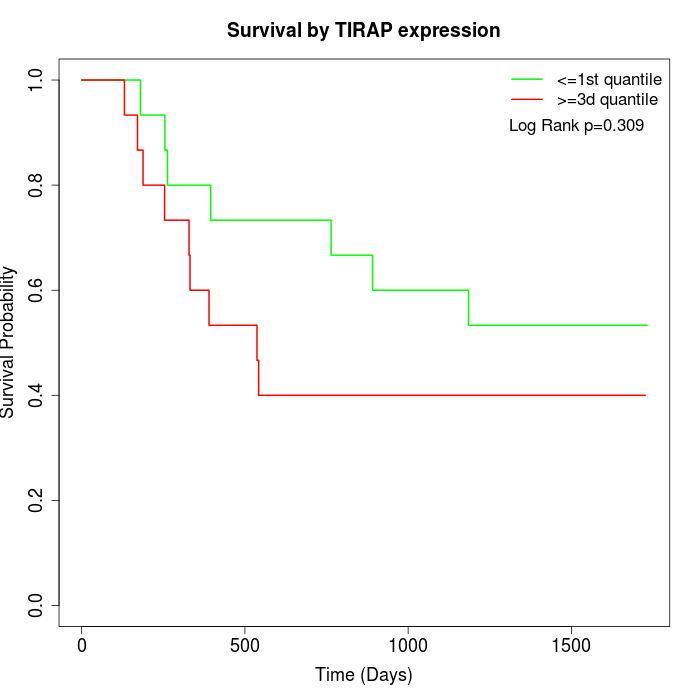
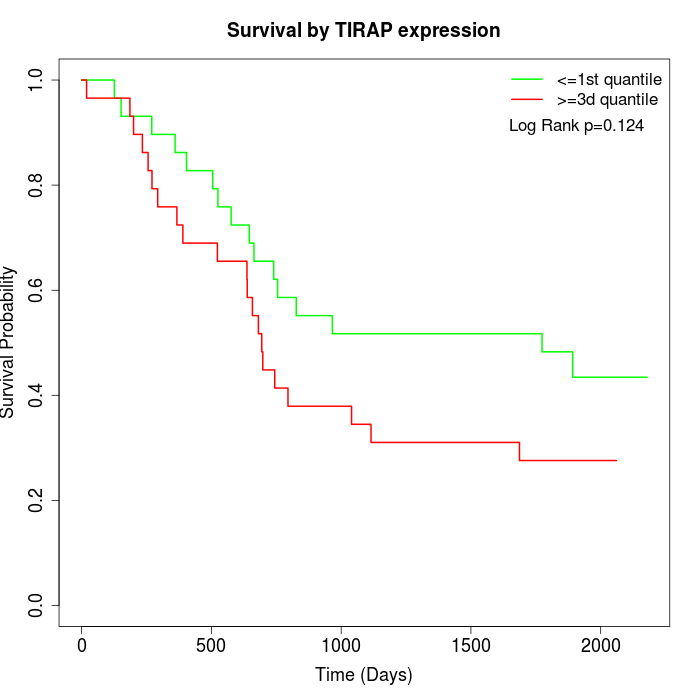
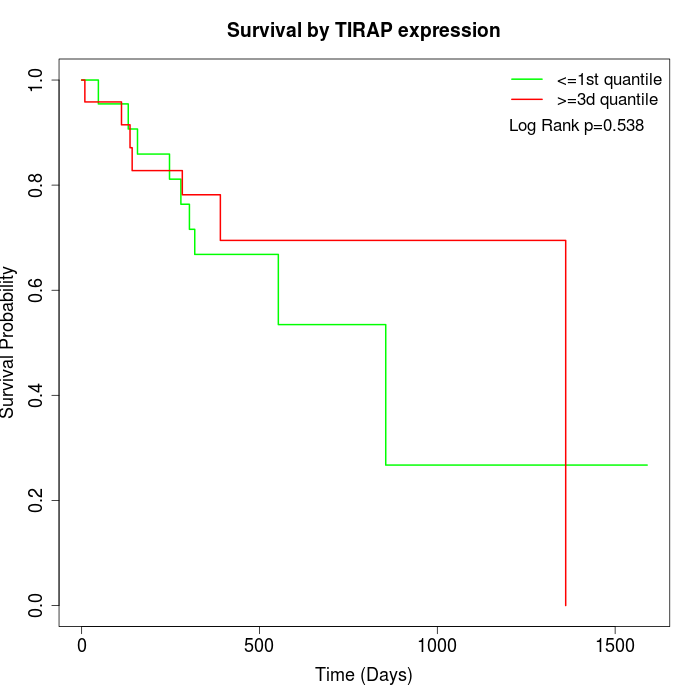
Survival by TIRAP expression:
Note: Click image to view full size file.
Copy number change of TIRAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TIRAP | 114609 | 0 | 13 | 17 | |
| GSE20123 | TIRAP | 114609 | 0 | 13 | 17 | |
| GSE43470 | TIRAP | 114609 | 2 | 7 | 34 | |
| GSE46452 | TIRAP | 114609 | 3 | 26 | 30 | |
| GSE47630 | TIRAP | 114609 | 2 | 19 | 19 | |
| GSE54993 | TIRAP | 114609 | 10 | 0 | 60 | |
| GSE54994 | TIRAP | 114609 | 5 | 19 | 29 | |
| GSE60625 | TIRAP | 114609 | 0 | 3 | 8 | |
| GSE74703 | TIRAP | 114609 | 1 | 5 | 30 | |
| GSE74704 | TIRAP | 114609 | 0 | 9 | 11 | |
| TCGA | TIRAP | 114609 | 7 | 50 | 39 |
Total number of gains: 30; Total number of losses: 164; Total Number of normals: 294.
Somatic mutations of TIRAP:
Generating mutation plots.
Highly correlated genes for TIRAP:
Showing top 20/219 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TIRAP | LYRM7 | 0.757257 | 3 | 0 | 3 |
| TIRAP | KCNK2 | 0.756723 | 3 | 0 | 3 |
| TIRAP | MRPS36 | 0.748542 | 3 | 0 | 3 |
| TIRAP | ZFP36L2 | 0.740348 | 3 | 0 | 3 |
| TIRAP | MPV17L | 0.737915 | 3 | 0 | 3 |
| TIRAP | ITPRIP | 0.734325 | 3 | 0 | 3 |
| TIRAP | RASSF4 | 0.720648 | 3 | 0 | 3 |
| TIRAP | TOM1L1 | 0.719312 | 4 | 0 | 3 |
| TIRAP | SNCA | 0.714494 | 3 | 0 | 3 |
| TIRAP | GNAT2 | 0.712753 | 3 | 0 | 3 |
| TIRAP | AMOTL2 | 0.707518 | 4 | 0 | 4 |
| TIRAP | FAM114A2 | 0.705693 | 3 | 0 | 3 |
| TIRAP | COBLL1 | 0.700936 | 3 | 0 | 3 |
| TIRAP | HOXA2 | 0.693259 | 4 | 0 | 4 |
| TIRAP | CETN3 | 0.690711 | 3 | 0 | 3 |
| TIRAP | ZNF540 | 0.689421 | 3 | 0 | 3 |
| TIRAP | KLF6 | 0.685683 | 3 | 0 | 3 |
| TIRAP | IL1RL1 | 0.677534 | 3 | 0 | 3 |
| TIRAP | FAM13C | 0.676764 | 3 | 0 | 3 |
| TIRAP | PRICKLE2 | 0.674087 | 4 | 0 | 3 |
For details and further investigation, click here