| Full name: phenylethanolamine N-methyltransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 5409 | HGNC ID: HGNC:9160 | Ensembl Gene: ENSG00000141744 | OMIM ID: 171190 |
| Drug and gene relationship at DGIdb | |||
Expression of PNMT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PNMT | 5409 | 206793_at | -0.1052 | 0.6053 | |
| GSE20347 | PNMT | 5409 | 206793_at | -0.0865 | 0.2894 | |
| GSE23400 | PNMT | 5409 | 206793_at | -0.1283 | 0.0001 | |
| GSE26886 | PNMT | 5409 | 206793_at | 0.1630 | 0.1416 | |
| GSE29001 | PNMT | 5409 | 206793_at | -0.0886 | 0.5797 | |
| GSE38129 | PNMT | 5409 | 206793_at | -0.1569 | 0.0317 | |
| GSE45670 | PNMT | 5409 | 206793_at | 0.0023 | 0.9832 | |
| GSE53622 | PNMT | 5409 | 94523 | -0.0554 | 0.7635 | |
| GSE53624 | PNMT | 5409 | 94523 | -0.5570 | 0.0003 | |
| GSE63941 | PNMT | 5409 | 206793_at | 0.2325 | 0.1673 | |
| GSE77861 | PNMT | 5409 | 206793_at | -0.1446 | 0.3061 | |
| TCGA | PNMT | 5409 | RNAseq | -1.7785 | 0.0098 |
Upregulated datasets: 0; Downregulated datasets: 1.
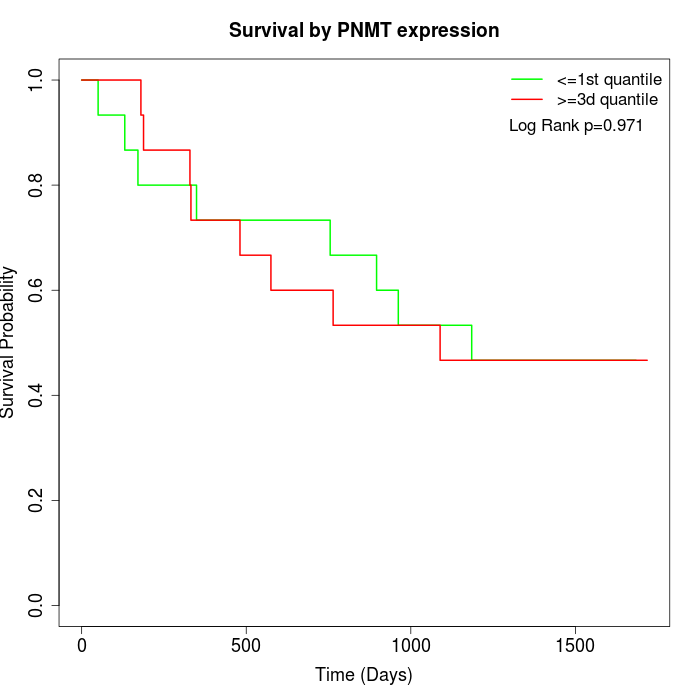
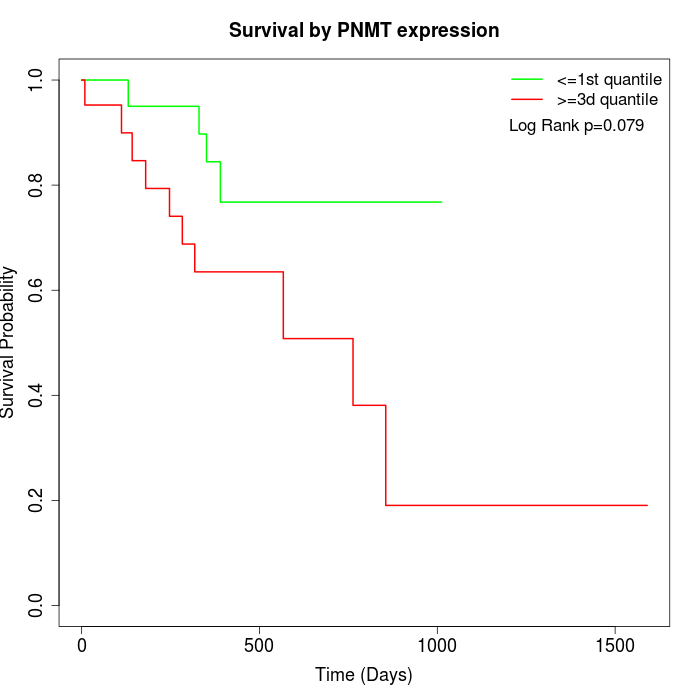
Survival by PNMT expression:
Note: Click image to view full size file.
Copy number change of PNMT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PNMT | 5409 | 8 | 1 | 21 | |
| GSE20123 | PNMT | 5409 | 8 | 1 | 21 | |
| GSE43470 | PNMT | 5409 | 1 | 1 | 41 | |
| GSE46452 | PNMT | 5409 | 34 | 0 | 25 | |
| GSE47630 | PNMT | 5409 | 8 | 1 | 31 | |
| GSE54993 | PNMT | 5409 | 3 | 4 | 63 | |
| GSE54994 | PNMT | 5409 | 9 | 5 | 39 | |
| GSE60625 | PNMT | 5409 | 4 | 0 | 7 | |
| GSE74703 | PNMT | 5409 | 1 | 0 | 35 | |
| GSE74704 | PNMT | 5409 | 5 | 1 | 14 | |
| TCGA | PNMT | 5409 | 23 | 8 | 65 |
Total number of gains: 104; Total number of losses: 22; Total Number of normals: 362.
Somatic mutations of PNMT:
Generating mutation plots.
Highly correlated genes for PNMT:
Showing top 20/648 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PNMT | RARA | 0.737631 | 4 | 0 | 4 |
| PNMT | KANK2 | 0.708983 | 3 | 0 | 3 |
| PNMT | CLRN3 | 0.705602 | 3 | 0 | 3 |
| PNMT | ABCC6 | 0.701343 | 3 | 0 | 3 |
| PNMT | ADAMTS7 | 0.697176 | 6 | 0 | 6 |
| PNMT | DNMT3L | 0.691365 | 5 | 0 | 5 |
| PNMT | DMRTA1 | 0.691129 | 3 | 0 | 3 |
| PNMT | KCND3 | 0.684543 | 5 | 0 | 4 |
| PNMT | NOTCH4 | 0.679031 | 3 | 0 | 3 |
| PNMT | GATA6 | 0.674429 | 3 | 0 | 3 |
| PNMT | DPPA4 | 0.673791 | 4 | 0 | 3 |
| PNMT | HMCN2 | 0.671059 | 3 | 0 | 3 |
| PNMT | GAP43 | 0.670131 | 4 | 0 | 4 |
| PNMT | CHRNA4 | 0.66961 | 6 | 0 | 5 |
| PNMT | SERPINA4 | 0.667934 | 4 | 0 | 4 |
| PNMT | SHANK1 | 0.663775 | 5 | 0 | 4 |
| PNMT | KCNF1 | 0.663085 | 6 | 0 | 5 |
| PNMT | CA4 | 0.657643 | 7 | 0 | 7 |
| PNMT | GRAP2 | 0.655805 | 4 | 0 | 4 |
| PNMT | DNAJC4 | 0.654785 | 3 | 0 | 3 |
For details and further investigation, click here