| Full name: thyroid hormone receptor interactor 6 | Alias Symbol: ZRP-1|OIP1|MGC10556|MGC10558|MGC29959|MGC3837|MGC4423 | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 7205 | HGNC ID: HGNC:12311 | Ensembl Gene: ENSG00000087077 | OMIM ID: 602933 |
| Drug and gene relationship at DGIdb | |||
TRIP6 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04621 | NOD-like receptor signaling pathway |
Expression of TRIP6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIP6 | 7205 | 209129_at | 0.4497 | 0.2213 | |
| GSE20347 | TRIP6 | 7205 | 209129_at | 0.2136 | 0.1908 | |
| GSE23400 | TRIP6 | 7205 | 209129_at | 0.2987 | 0.0013 | |
| GSE26886 | TRIP6 | 7205 | 209129_at | -0.2534 | 0.3568 | |
| GSE29001 | TRIP6 | 7205 | 209129_at | 0.3113 | 0.5230 | |
| GSE38129 | TRIP6 | 7205 | 209129_at | 0.2601 | 0.0925 | |
| GSE45670 | TRIP6 | 7205 | 209129_at | 0.2616 | 0.1726 | |
| GSE53622 | TRIP6 | 7205 | 30297 | 0.2828 | 0.0005 | |
| GSE53624 | TRIP6 | 7205 | 30297 | 0.2343 | 0.0002 | |
| GSE63941 | TRIP6 | 7205 | 209129_at | -0.0090 | 0.9943 | |
| GSE77861 | TRIP6 | 7205 | 209129_at | 0.0981 | 0.6399 | |
| GSE97050 | TRIP6 | 7205 | A_24_P4054 | -0.0910 | 0.7724 | |
| SRP007169 | TRIP6 | 7205 | RNAseq | -0.2968 | 0.4218 | |
| SRP008496 | TRIP6 | 7205 | RNAseq | -0.0612 | 0.8207 | |
| SRP064894 | TRIP6 | 7205 | RNAseq | 0.1204 | 0.5946 | |
| SRP133303 | TRIP6 | 7205 | RNAseq | 0.0654 | 0.7666 | |
| SRP159526 | TRIP6 | 7205 | RNAseq | -0.1460 | 0.6227 | |
| SRP193095 | TRIP6 | 7205 | RNAseq | 0.2412 | 0.2120 | |
| SRP219564 | TRIP6 | 7205 | RNAseq | -0.1446 | 0.7311 | |
| TCGA | TRIP6 | 7205 | RNAseq | 0.3888 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
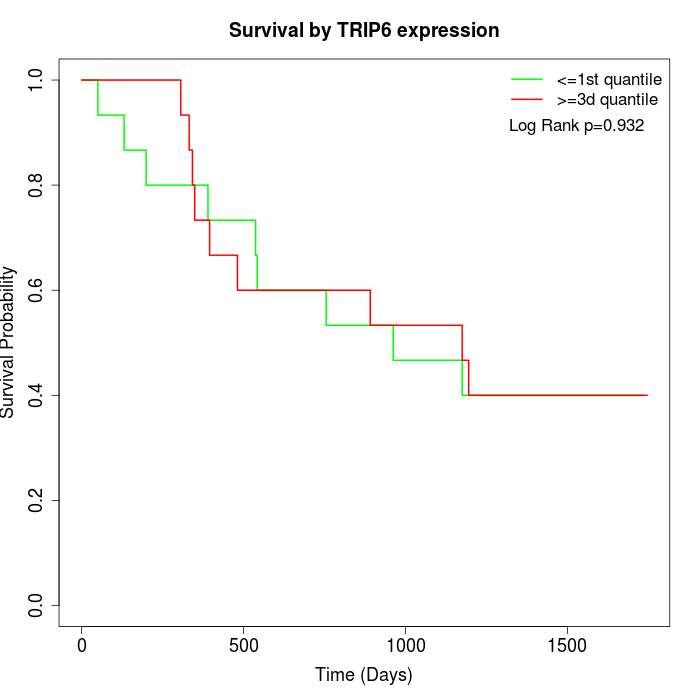
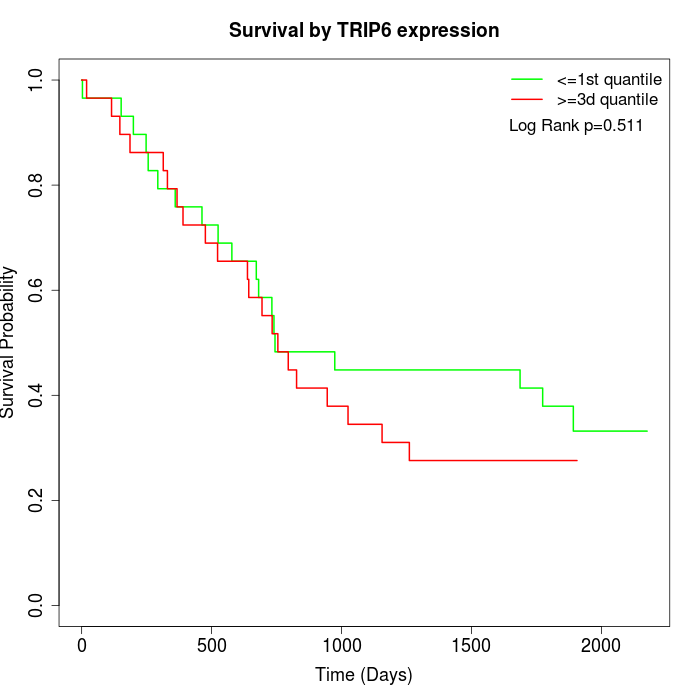
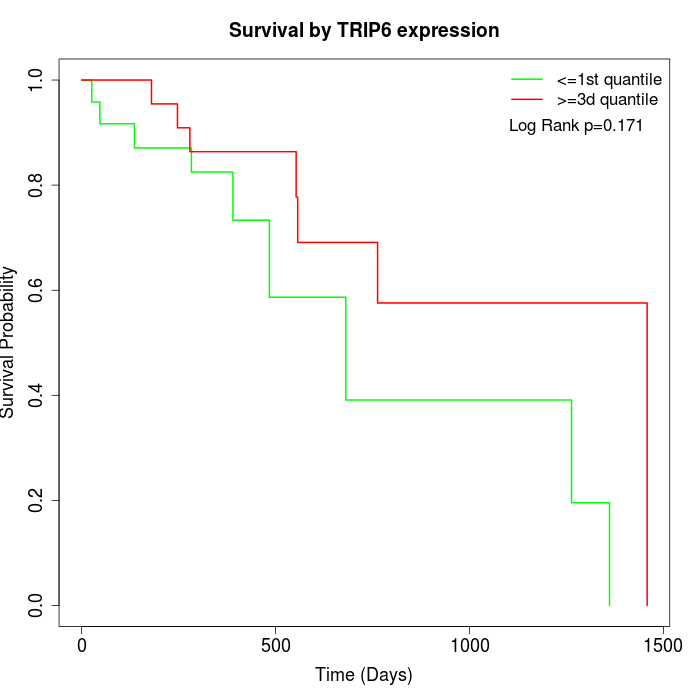
Survival by TRIP6 expression:
Note: Click image to view full size file.
Copy number change of TRIP6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIP6 | 7205 | 13 | 0 | 17 | |
| GSE20123 | TRIP6 | 7205 | 13 | 0 | 17 | |
| GSE43470 | TRIP6 | 7205 | 7 | 2 | 34 | |
| GSE46452 | TRIP6 | 7205 | 11 | 1 | 47 | |
| GSE47630 | TRIP6 | 7205 | 7 | 3 | 30 | |
| GSE54993 | TRIP6 | 7205 | 1 | 9 | 60 | |
| GSE54994 | TRIP6 | 7205 | 15 | 3 | 35 | |
| GSE60625 | TRIP6 | 7205 | 0 | 0 | 11 | |
| GSE74703 | TRIP6 | 7205 | 7 | 1 | 28 | |
| GSE74704 | TRIP6 | 7205 | 9 | 0 | 11 | |
| TCGA | TRIP6 | 7205 | 52 | 7 | 37 |
Total number of gains: 135; Total number of losses: 26; Total Number of normals: 327.
Somatic mutations of TRIP6:
Generating mutation plots.
Highly correlated genes for TRIP6:
Showing top 20/347 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIP6 | ATXN7L3B | 0.73508 | 3 | 0 | 3 |
| TRIP6 | PRDM15 | 0.730322 | 3 | 0 | 3 |
| TRIP6 | GCDH | 0.725485 | 3 | 0 | 3 |
| TRIP6 | MAP3K13 | 0.723184 | 3 | 0 | 3 |
| TRIP6 | CCNT1 | 0.720927 | 3 | 0 | 3 |
| TRIP6 | RNF214 | 0.718266 | 3 | 0 | 3 |
| TRIP6 | SSBP4 | 0.70873 | 3 | 0 | 3 |
| TRIP6 | PPIE | 0.703747 | 3 | 0 | 3 |
| TRIP6 | FAM89B | 0.698814 | 3 | 0 | 3 |
| TRIP6 | LRRC58 | 0.695919 | 3 | 0 | 3 |
| TRIP6 | MRI1 | 0.694651 | 3 | 0 | 3 |
| TRIP6 | ARHGEF11 | 0.688936 | 3 | 0 | 3 |
| TRIP6 | RNF113A | 0.688774 | 3 | 0 | 3 |
| TRIP6 | PRR3 | 0.687275 | 4 | 0 | 3 |
| TRIP6 | SLC25A19 | 0.682125 | 3 | 0 | 3 |
| TRIP6 | FAM193A | 0.672722 | 3 | 0 | 3 |
| TRIP6 | SENP5 | 0.671937 | 4 | 0 | 3 |
| TRIP6 | THADA | 0.669342 | 3 | 0 | 3 |
| TRIP6 | TUBA1B | 0.667051 | 4 | 0 | 3 |
| TRIP6 | SMARCC1 | 0.66658 | 4 | 0 | 3 |
For details and further investigation, click here