| Full name: transient receptor potential cation channel subfamily M member 5 | Alias Symbol: LTRPC5|MTR1 | ||
| Type: protein-coding gene | Cytoband: 11p15.5 | ||
| Entrez ID: 29850 | HGNC ID: HGNC:14323 | Ensembl Gene: ENSG00000070985 | OMIM ID: 604600 |
| Drug and gene relationship at DGIdb | |||
TRPM5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04742 | Taste transduction |
Expression of TRPM5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRPM5 | 29850 | 223935_at | 0.1520 | 0.3901 | |
| GSE26886 | TRPM5 | 29850 | 223935_at | 0.3088 | 0.0095 | |
| GSE45670 | TRPM5 | 29850 | 223935_at | 0.0696 | 0.4705 | |
| GSE53622 | TRPM5 | 29850 | 56898 | 0.1003 | 0.1457 | |
| GSE53624 | TRPM5 | 29850 | 56898 | -0.0409 | 0.7395 | |
| GSE63941 | TRPM5 | 29850 | 223935_at | 0.2595 | 0.0981 | |
| GSE77861 | TRPM5 | 29850 | 223935_at | -0.1173 | 0.1937 | |
| TCGA | TRPM5 | 29850 | RNAseq | -2.1363 | 0.0493 |
Upregulated datasets: 0; Downregulated datasets: 1.
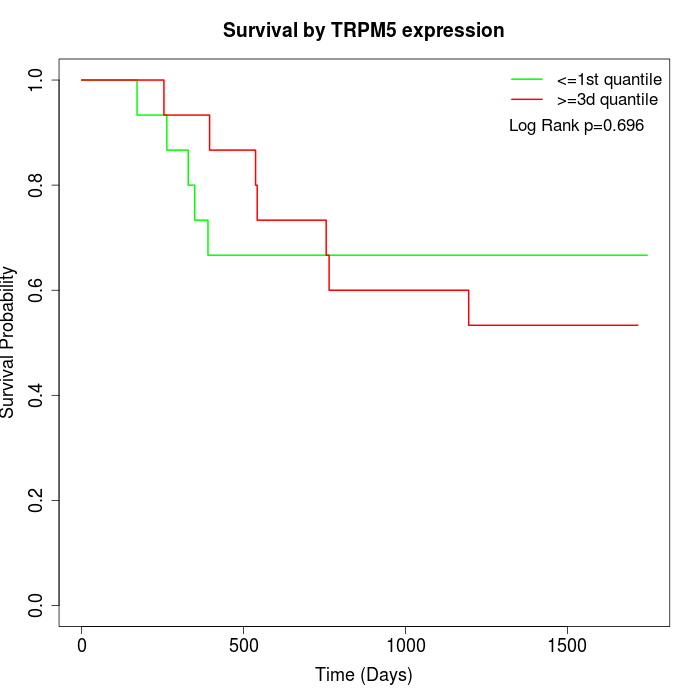
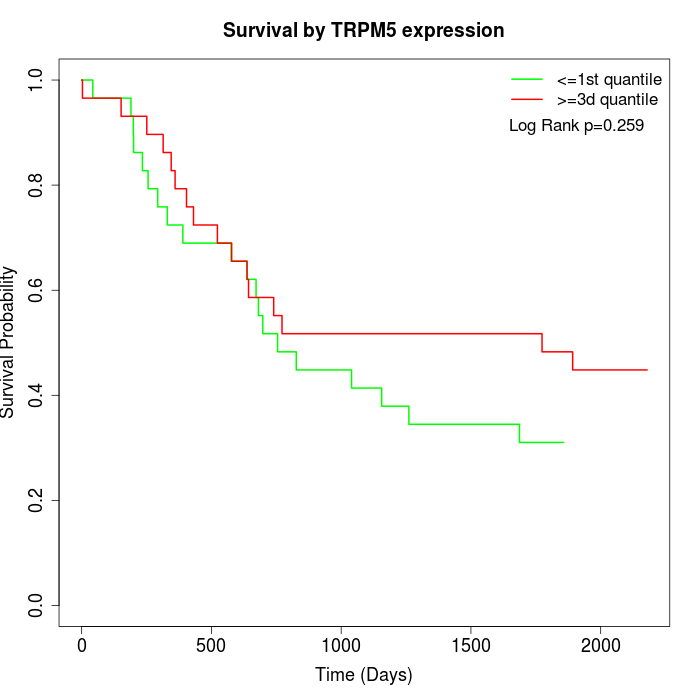
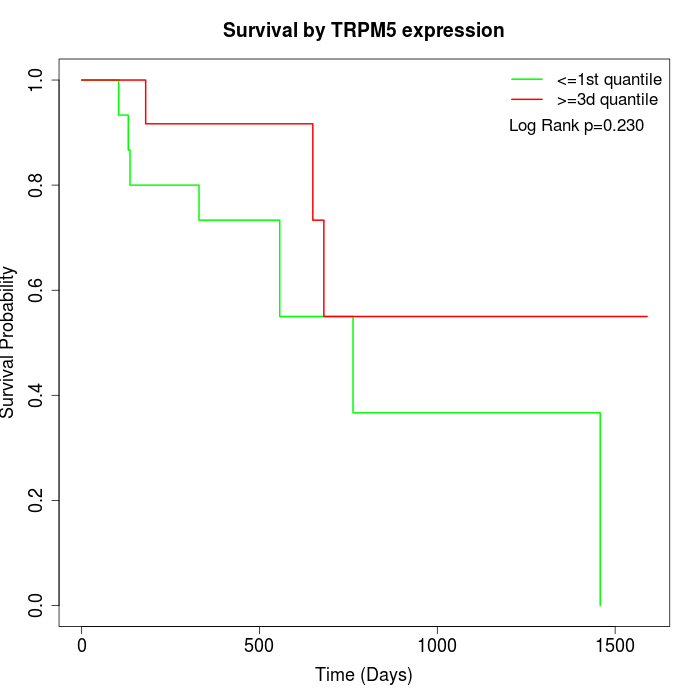
Survival by TRPM5 expression:
Note: Click image to view full size file.
Copy number change of TRPM5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRPM5 | 29850 | 1 | 11 | 18 | |
| GSE20123 | TRPM5 | 29850 | 1 | 12 | 17 | |
| GSE43470 | TRPM5 | 29850 | 1 | 9 | 33 | |
| GSE46452 | TRPM5 | 29850 | 7 | 8 | 44 | |
| GSE47630 | TRPM5 | 29850 | 4 | 12 | 24 | |
| GSE54993 | TRPM5 | 29850 | 3 | 1 | 66 | |
| GSE54994 | TRPM5 | 29850 | 1 | 12 | 40 | |
| GSE60625 | TRPM5 | 29850 | 0 | 0 | 11 | |
| GSE74703 | TRPM5 | 29850 | 1 | 7 | 28 | |
| GSE74704 | TRPM5 | 29850 | 0 | 8 | 12 | |
| TCGA | TRPM5 | 29850 | 8 | 39 | 49 |
Total number of gains: 27; Total number of losses: 119; Total Number of normals: 342.
Somatic mutations of TRPM5:
Generating mutation plots.
Highly correlated genes for TRPM5:
Showing top 20/177 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRPM5 | GRIK4 | 0.727044 | 3 | 0 | 3 |
| TRPM5 | SERPINA10 | 0.692335 | 4 | 0 | 4 |
| TRPM5 | IMPG2 | 0.682195 | 3 | 0 | 3 |
| TRPM5 | BDNF-AS | 0.667188 | 3 | 0 | 3 |
| TRPM5 | CCIN | 0.666277 | 3 | 0 | 3 |
| TRPM5 | SLC22A31 | 0.665871 | 3 | 0 | 3 |
| TRPM5 | INSL3 | 0.656163 | 3 | 0 | 3 |
| TRPM5 | TNFRSF9 | 0.654167 | 3 | 0 | 3 |
| TRPM5 | PRDM13 | 0.651797 | 3 | 0 | 3 |
| TRPM5 | UPK3A | 0.651571 | 4 | 0 | 4 |
| TRPM5 | NANOS3 | 0.648076 | 3 | 0 | 3 |
| TRPM5 | SLC5A12 | 0.643776 | 3 | 0 | 3 |
| TRPM5 | MYOD1 | 0.641801 | 3 | 0 | 3 |
| TRPM5 | ZSCAN2 | 0.638201 | 3 | 0 | 3 |
| TRPM5 | NPR1 | 0.634924 | 3 | 0 | 3 |
| TRPM5 | CLDN9 | 0.634269 | 4 | 0 | 4 |
| TRPM5 | FZD9 | 0.632021 | 4 | 0 | 4 |
| TRPM5 | TTLL9 | 0.631979 | 3 | 0 | 3 |
| TRPM5 | PRDM8 | 0.626834 | 3 | 0 | 3 |
| TRPM5 | GPC2 | 0.626497 | 3 | 0 | 3 |
For details and further investigation, click here