| Full name: vasoactive intestinal peptide | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6q25.2 | ||
| Entrez ID: 7432 | HGNC ID: HGNC:12693 | Ensembl Gene: ENSG00000146469 | OMIM ID: 192320 |
| Drug and gene relationship at DGIdb | |||
Expression of VIP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VIP | 7432 | 206577_at | -0.7510 | 0.6496 | |
| GSE20347 | VIP | 7432 | 206577_at | -0.2257 | 0.3005 | |
| GSE23400 | VIP | 7432 | 206577_at | -0.1123 | 0.0062 | |
| GSE26886 | VIP | 7432 | 206577_at | -0.1814 | 0.0353 | |
| GSE29001 | VIP | 7432 | 206577_at | -0.2636 | 0.0365 | |
| GSE38129 | VIP | 7432 | 206577_at | -0.8372 | 0.0256 | |
| GSE45670 | VIP | 7432 | 206577_at | -0.2429 | 0.0429 | |
| GSE53622 | VIP | 7432 | 65072 | -0.7128 | 0.0307 | |
| GSE53624 | VIP | 7432 | 65072 | -0.2791 | 0.1046 | |
| GSE63941 | VIP | 7432 | 206577_at | 0.0674 | 0.6416 | |
| GSE77861 | VIP | 7432 | 206577_at | -0.0889 | 0.2409 | |
| GSE97050 | VIP | 7432 | A_23_P19650 | -0.8036 | 0.1998 | |
| TCGA | VIP | 7432 | RNAseq | -2.7409 | 0.0006 |
Upregulated datasets: 0; Downregulated datasets: 1.
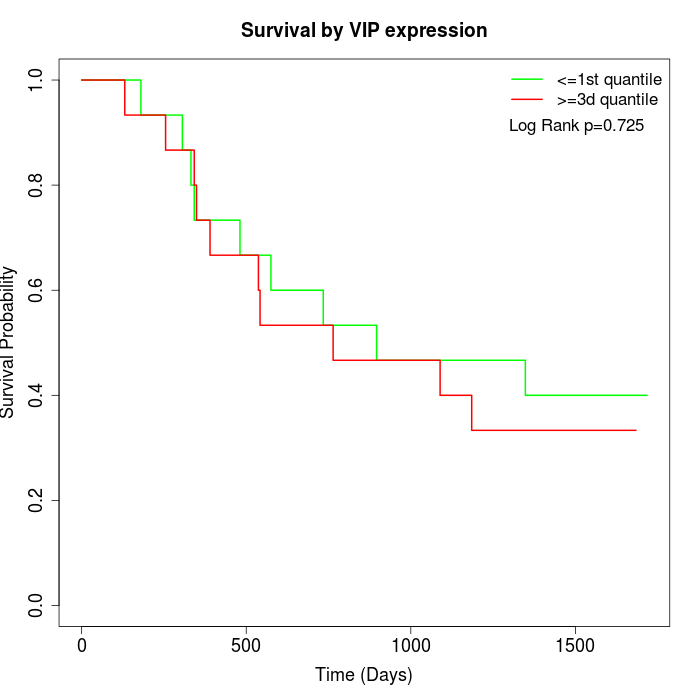
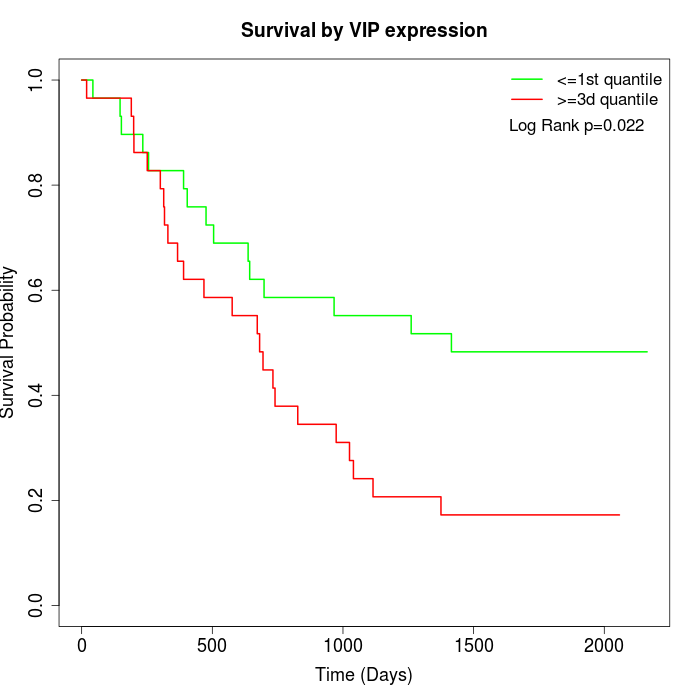
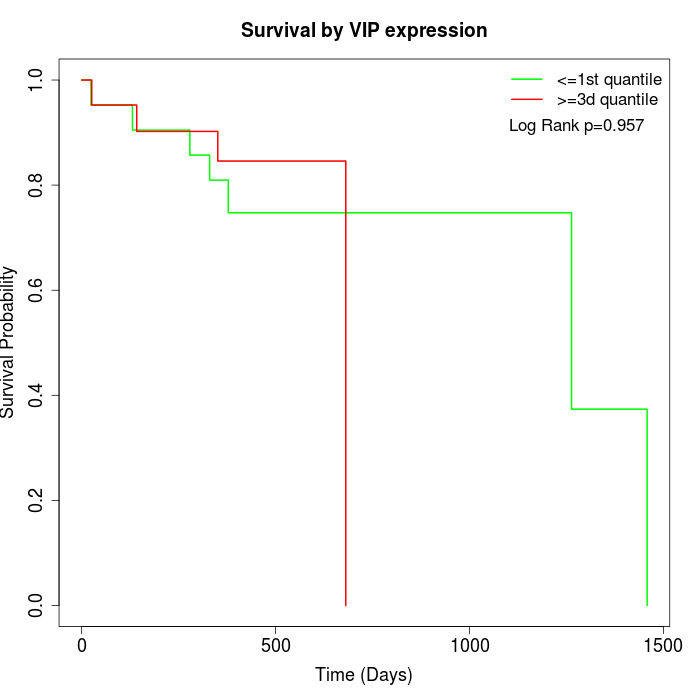
Survival by VIP expression:
Note: Click image to view full size file.
Copy number change of VIP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VIP | 7432 | 1 | 4 | 25 | |
| GSE20123 | VIP | 7432 | 1 | 3 | 26 | |
| GSE43470 | VIP | 7432 | 4 | 0 | 39 | |
| GSE46452 | VIP | 7432 | 3 | 10 | 46 | |
| GSE47630 | VIP | 7432 | 9 | 4 | 27 | |
| GSE54993 | VIP | 7432 | 3 | 2 | 65 | |
| GSE54994 | VIP | 7432 | 8 | 8 | 37 | |
| GSE60625 | VIP | 7432 | 0 | 1 | 10 | |
| GSE74703 | VIP | 7432 | 4 | 0 | 32 | |
| GSE74704 | VIP | 7432 | 0 | 1 | 19 | |
| TCGA | VIP | 7432 | 11 | 21 | 64 |
Total number of gains: 44; Total number of losses: 54; Total Number of normals: 390.
Somatic mutations of VIP:
Generating mutation plots.
Highly correlated genes for VIP:
Showing top 20/458 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VIP | SLC35F1 | 0.775221 | 3 | 0 | 3 |
| VIP | GIPC3 | 0.773787 | 3 | 0 | 3 |
| VIP | GPM6A | 0.75202 | 7 | 0 | 7 |
| VIP | SH3GL2 | 0.734991 | 7 | 0 | 7 |
| VIP | ESRRG | 0.729605 | 5 | 0 | 5 |
| VIP | NPY | 0.727414 | 10 | 0 | 8 |
| VIP | IQUB | 0.723442 | 4 | 0 | 3 |
| VIP | RYR2 | 0.722566 | 6 | 0 | 5 |
| VIP | DCLK2 | 0.722385 | 5 | 0 | 4 |
| VIP | LRRTM1 | 0.71803 | 4 | 0 | 3 |
| VIP | BRINP3 | 0.715237 | 6 | 0 | 6 |
| VIP | FBXO30 | 0.707879 | 3 | 0 | 3 |
| VIP | TMOD1 | 0.706836 | 6 | 0 | 5 |
| VIP | OMP | 0.704042 | 3 | 0 | 3 |
| VIP | SYT4 | 0.70143 | 3 | 0 | 3 |
| VIP | ARHGAP36 | 0.693732 | 3 | 0 | 3 |
| VIP | ENOX1 | 0.691784 | 6 | 0 | 6 |
| VIP | BHMT2 | 0.689696 | 8 | 0 | 7 |
| VIP | LMO3 | 0.68947 | 9 | 0 | 7 |
| VIP | CARTPT | 0.689412 | 7 | 0 | 6 |
For details and further investigation, click here