| Full name: maltase-glucoamylase | Alias Symbol: MGA | ||
| Type: protein-coding gene | Cytoband: 7q34 | ||
| Entrez ID: 8972 | HGNC ID: HGNC:7043 | Ensembl Gene: ENSG00000257335 | OMIM ID: 154360 |
| Drug and gene relationship at DGIdb | |||
Expression of MGAM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MGAM | 8972 | 206522_at | 0.8143 | 0.1718 | |
| GSE20347 | MGAM | 8972 | 206522_at | 0.0544 | 0.2990 | |
| GSE23400 | MGAM | 8972 | 206522_at | -0.0165 | 0.3850 | |
| GSE26886 | MGAM | 8972 | 206522_at | 0.1713 | 0.2726 | |
| GSE29001 | MGAM | 8972 | 206522_at | 0.1727 | 0.2508 | |
| GSE38129 | MGAM | 8972 | 206522_at | 0.0195 | 0.8152 | |
| GSE45670 | MGAM | 8972 | 206522_at | 0.1477 | 0.5965 | |
| GSE53622 | MGAM | 8972 | 96189 | -0.0579 | 0.8319 | |
| GSE53624 | MGAM | 8972 | 136615 | 0.3214 | 0.0000 | |
| GSE63941 | MGAM | 8972 | 206522_at | 0.3935 | 0.5809 | |
| GSE77861 | MGAM | 8972 | 206522_at | -0.0369 | 0.7425 | |
| GSE97050 | MGAM | 8972 | A_33_P3409765 | -0.3989 | 0.5934 | |
| SRP133303 | MGAM | 8972 | RNAseq | 2.0918 | 0.0000 | |
| SRP159526 | MGAM | 8972 | RNAseq | 4.1431 | 0.0000 | |
| SRP193095 | MGAM | 8972 | RNAseq | -0.3489 | 0.5265 | |
| TCGA | MGAM | 8972 | RNAseq | -1.6352 | 0.0061 |
Upregulated datasets: 2; Downregulated datasets: 1.
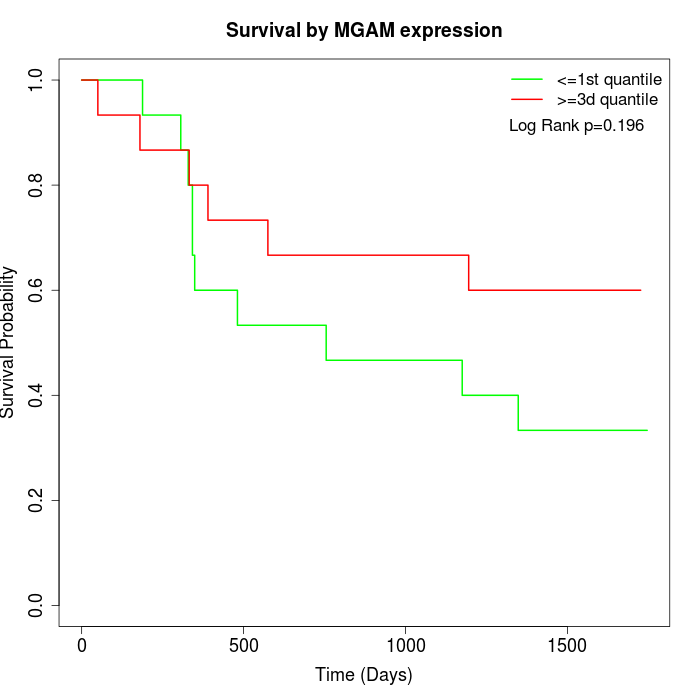
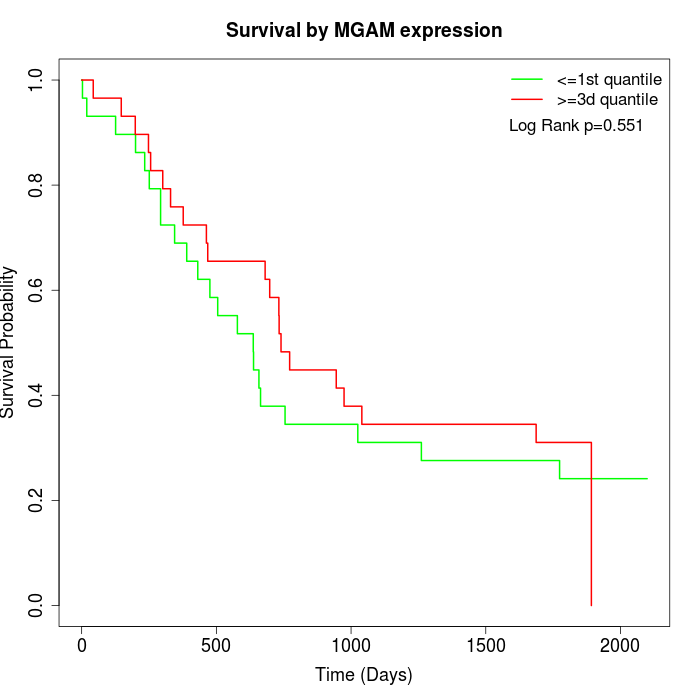
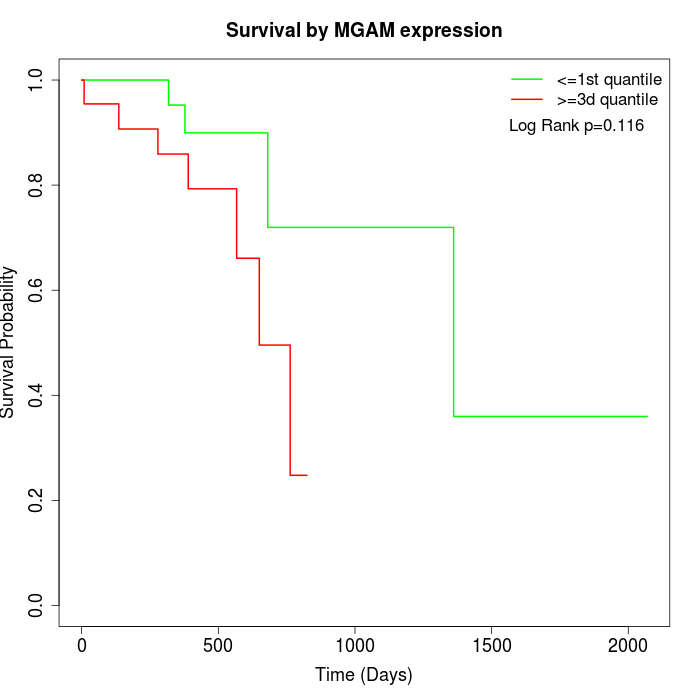
Survival by MGAM expression:
Note: Click image to view full size file.
Copy number change of MGAM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MGAM | 8972 | 5 | 2 | 23 | |
| GSE20123 | MGAM | 8972 | 5 | 2 | 23 | |
| GSE43470 | MGAM | 8972 | 2 | 5 | 36 | |
| GSE46452 | MGAM | 8972 | 7 | 2 | 50 | |
| GSE47630 | MGAM | 8972 | 6 | 7 | 27 | |
| GSE54993 | MGAM | 8972 | 4 | 4 | 62 | |
| GSE54994 | MGAM | 8972 | 5 | 8 | 40 | |
| GSE60625 | MGAM | 8972 | 0 | 0 | 11 | |
| GSE74703 | MGAM | 8972 | 2 | 4 | 30 | |
| GSE74704 | MGAM | 8972 | 1 | 2 | 17 | |
| TCGA | MGAM | 8972 | 29 | 23 | 44 |
Total number of gains: 66; Total number of losses: 59; Total Number of normals: 363.
Somatic mutations of MGAM:
Generating mutation plots.
Highly correlated genes for MGAM:
Showing top 20/37 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MGAM | WFDC3 | 0.769367 | 3 | 0 | 3 |
| MGAM | DDC | 0.717287 | 3 | 0 | 3 |
| MGAM | CMTM2 | 0.653957 | 6 | 0 | 5 |
| MGAM | LILRA3 | 0.638776 | 4 | 0 | 3 |
| MGAM | HUNK | 0.615882 | 3 | 0 | 3 |
| MGAM | TNFRSF10C | 0.601193 | 3 | 0 | 3 |
| MGAM | ARC | 0.598056 | 4 | 0 | 3 |
| MGAM | VNN2 | 0.596842 | 8 | 0 | 5 |
| MGAM | VMO1 | 0.585175 | 4 | 0 | 3 |
| MGAM | TMEM8B | 0.58349 | 4 | 0 | 3 |
| MGAM | CCDC177 | 0.582287 | 3 | 0 | 3 |
| MGAM | PROK2 | 0.577695 | 6 | 0 | 5 |
| MGAM | RGS16 | 0.577283 | 4 | 0 | 3 |
| MGAM | FGF9 | 0.571466 | 3 | 0 | 3 |
| MGAM | TRHDE | 0.5657 | 3 | 0 | 3 |
| MGAM | CXCR1 | 0.561329 | 7 | 0 | 4 |
| MGAM | SIGLEC1 | 0.559157 | 3 | 0 | 3 |
| MGAM | FRS3 | 0.553505 | 4 | 0 | 3 |
| MGAM | SCUBE3 | 0.545362 | 4 | 0 | 3 |
| MGAM | C5AR1 | 0.542775 | 7 | 0 | 5 |
For details and further investigation, click here