| Full name: WNK lysine deficient protein kinase 2 | Alias Symbol: NY-CO-43|KIAA1760 | ||
| Type: protein-coding gene | Cytoband: 9q22.31 | ||
| Entrez ID: 65268 | HGNC ID: HGNC:14542 | Ensembl Gene: ENSG00000165238 | OMIM ID: 606249 |
| Drug and gene relationship at DGIdb | |||
Expression of WNK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | WNK2 | 65268 | 227217_at | 0.0641 | 0.9508 | |
| GSE26886 | WNK2 | 65268 | 1560866_at | -0.2717 | 0.0164 | |
| GSE45670 | WNK2 | 65268 | 1557536_at | 0.1494 | 0.0752 | |
| GSE53622 | WNK2 | 65268 | 71839 | -0.1662 | 0.2307 | |
| GSE53624 | WNK2 | 65268 | 25169 | -0.2592 | 0.2150 | |
| GSE63941 | WNK2 | 65268 | 1562026_at | 0.3498 | 0.0481 | |
| GSE77861 | WNK2 | 65268 | 1560866_at | -0.2919 | 0.0018 | |
| GSE97050 | WNK2 | 65268 | A_33_P3305885 | -0.9324 | 0.2199 | |
| SRP007169 | WNK2 | 65268 | RNAseq | 6.4909 | 0.0000 | |
| SRP064894 | WNK2 | 65268 | RNAseq | -0.1859 | 0.6860 | |
| SRP133303 | WNK2 | 65268 | RNAseq | 0.1384 | 0.8223 | |
| SRP159526 | WNK2 | 65268 | RNAseq | 1.9975 | 0.0000 | |
| SRP193095 | WNK2 | 65268 | RNAseq | 1.3761 | 0.0004 | |
| SRP219564 | WNK2 | 65268 | RNAseq | -0.0895 | 0.9351 | |
| TCGA | WNK2 | 65268 | RNAseq | -0.3741 | 0.0833 |
Upregulated datasets: 3; Downregulated datasets: 0.
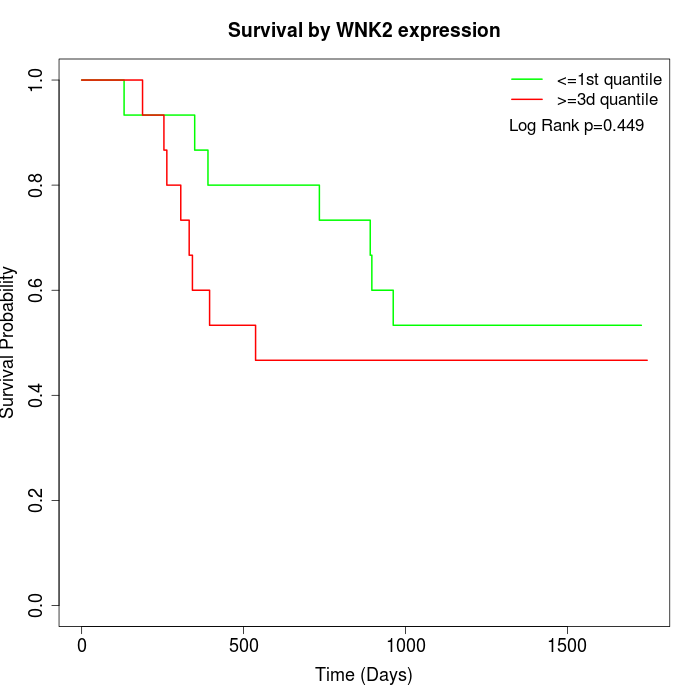
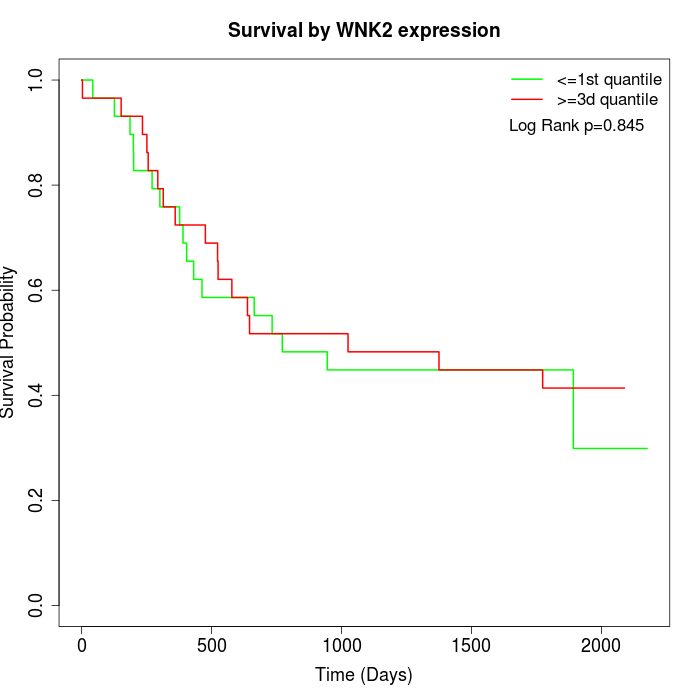
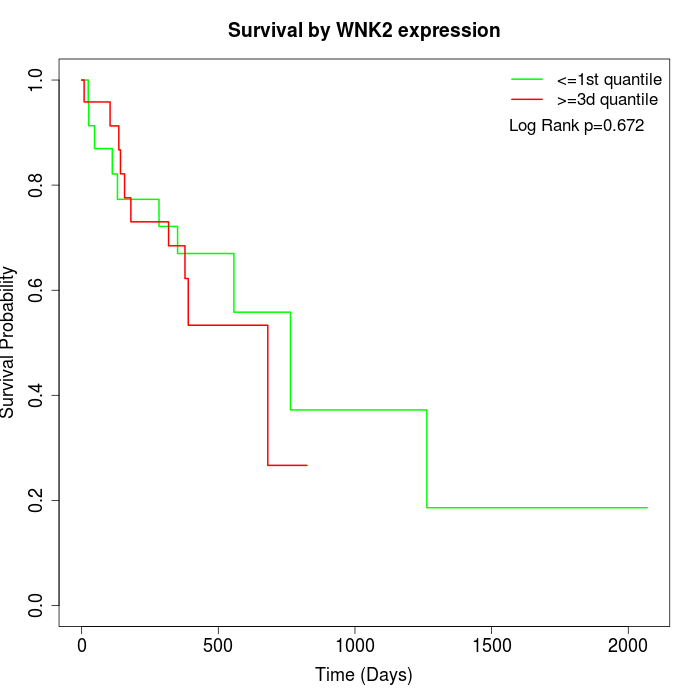
Survival by WNK2 expression:
Note: Click image to view full size file.
Copy number change of WNK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | WNK2 | 65268 | 5 | 9 | 16 | |
| GSE20123 | WNK2 | 65268 | 5 | 9 | 16 | |
| GSE43470 | WNK2 | 65268 | 6 | 3 | 34 | |
| GSE46452 | WNK2 | 65268 | 6 | 14 | 39 | |
| GSE47630 | WNK2 | 65268 | 1 | 17 | 22 | |
| GSE54993 | WNK2 | 65268 | 4 | 2 | 64 | |
| GSE54994 | WNK2 | 65268 | 8 | 11 | 34 | |
| GSE60625 | WNK2 | 65268 | 0 | 0 | 11 | |
| GSE74703 | WNK2 | 65268 | 5 | 3 | 28 | |
| GSE74704 | WNK2 | 65268 | 2 | 7 | 11 | |
| TCGA | WNK2 | 65268 | 24 | 23 | 49 |
Total number of gains: 66; Total number of losses: 98; Total Number of normals: 324.
Somatic mutations of WNK2:
Generating mutation plots.
Highly correlated genes for WNK2:
Showing top 20/239 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| WNK2 | CLDN3 | 0.764709 | 3 | 0 | 3 |
| WNK2 | ODF4 | 0.740306 | 3 | 0 | 3 |
| WNK2 | ZNF280A | 0.730978 | 3 | 0 | 3 |
| WNK2 | SLC25A34 | 0.707297 | 3 | 0 | 3 |
| WNK2 | HTR1E | 0.701413 | 3 | 0 | 3 |
| WNK2 | DSCR8 | 0.697724 | 3 | 0 | 3 |
| WNK2 | ST3GAL3 | 0.691757 | 4 | 0 | 3 |
| WNK2 | DNAJB1 | 0.691054 | 3 | 0 | 3 |
| WNK2 | TNNT3 | 0.688695 | 4 | 0 | 3 |
| WNK2 | PPL | 0.688308 | 3 | 0 | 3 |
| WNK2 | ABCC8 | 0.681703 | 4 | 0 | 3 |
| WNK2 | LCN8 | 0.67492 | 3 | 0 | 3 |
| WNK2 | CA1 | 0.673626 | 4 | 0 | 3 |
| WNK2 | LRRC43 | 0.672699 | 3 | 0 | 3 |
| WNK2 | SNORA71A | 0.671428 | 3 | 0 | 3 |
| WNK2 | FRMD1 | 0.67094 | 3 | 0 | 3 |
| WNK2 | DEPTOR | 0.668098 | 3 | 0 | 3 |
| WNK2 | CRTAC1 | 0.667765 | 3 | 0 | 3 |
| WNK2 | APOM | 0.667299 | 3 | 0 | 3 |
| WNK2 | MCRS1 | 0.666838 | 3 | 0 | 3 |
For details and further investigation, click here