| Full name: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 7529 | HGNC ID: HGNC:12849 | Ensembl Gene: ENSG00000166913 | OMIM ID: 601289 |
| Drug and gene relationship at DGIdb | |||
YWHAB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04390 | Hippo signaling pathway |
Expression of YWHAB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | YWHAB | 7529 | 217718_s_at | 0.2018 | 0.6337 | |
| GSE20347 | YWHAB | 7529 | 217718_s_at | -0.3805 | 0.0002 | |
| GSE23400 | YWHAB | 7529 | 217718_s_at | -0.0333 | 0.5684 | |
| GSE26886 | YWHAB | 7529 | 217718_s_at | -0.0137 | 0.9441 | |
| GSE29001 | YWHAB | 7529 | 217718_s_at | -0.0006 | 0.9957 | |
| GSE38129 | YWHAB | 7529 | 217718_s_at | -0.1696 | 0.0602 | |
| GSE45670 | YWHAB | 7529 | 217718_s_at | 0.0003 | 0.9977 | |
| GSE53622 | YWHAB | 7529 | 40277 | -0.4191 | 0.0000 | |
| GSE53624 | YWHAB | 7529 | 40277 | -0.2988 | 0.0000 | |
| GSE63941 | YWHAB | 7529 | 217718_s_at | -0.3917 | 0.3057 | |
| GSE77861 | YWHAB | 7529 | 217718_s_at | -0.2646 | 0.0693 | |
| GSE97050 | YWHAB | 7529 | A_23_P120414 | -0.3122 | 0.2699 | |
| SRP007169 | YWHAB | 7529 | RNAseq | -1.3299 | 0.0000 | |
| SRP008496 | YWHAB | 7529 | RNAseq | -1.0535 | 0.0000 | |
| SRP064894 | YWHAB | 7529 | RNAseq | -0.1598 | 0.1803 | |
| SRP133303 | YWHAB | 7529 | RNAseq | -0.0373 | 0.6739 | |
| SRP159526 | YWHAB | 7529 | RNAseq | -0.4274 | 0.0108 | |
| SRP193095 | YWHAB | 7529 | RNAseq | -0.5112 | 0.0000 | |
| SRP219564 | YWHAB | 7529 | RNAseq | -0.0950 | 0.6658 | |
| TCGA | YWHAB | 7529 | RNAseq | 0.0663 | 0.0743 |
Upregulated datasets: 0; Downregulated datasets: 2.
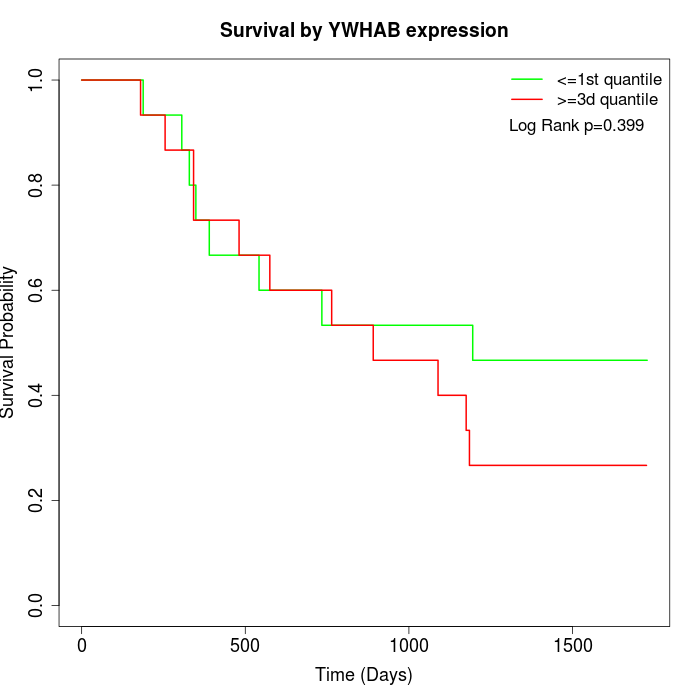
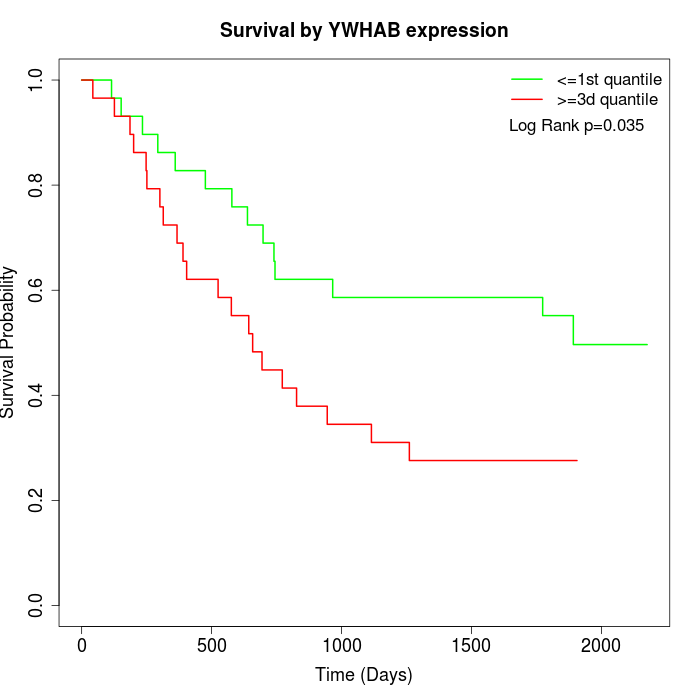
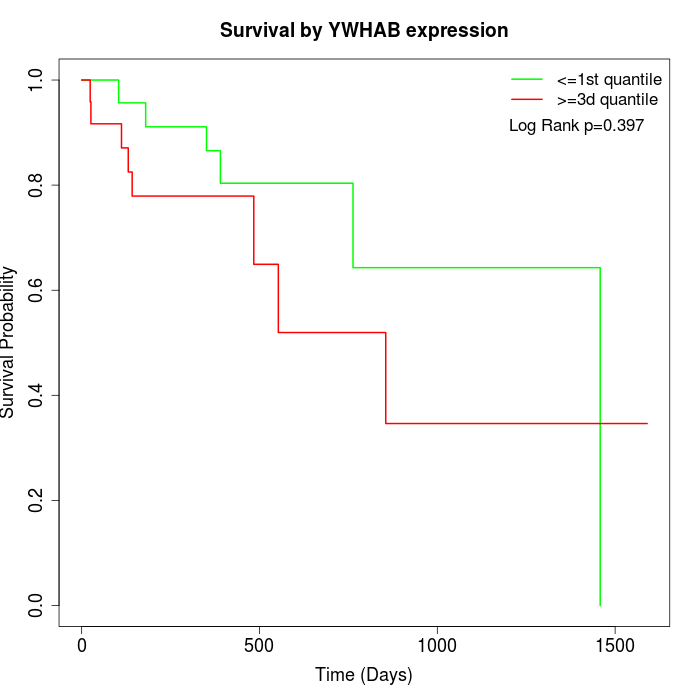
Survival by YWHAB expression:
Note: Click image to view full size file.
Copy number change of YWHAB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | YWHAB | 7529 | 13 | 1 | 16 | |
| GSE20123 | YWHAB | 7529 | 13 | 1 | 16 | |
| GSE43470 | YWHAB | 7529 | 12 | 1 | 30 | |
| GSE46452 | YWHAB | 7529 | 29 | 0 | 30 | |
| GSE47630 | YWHAB | 7529 | 24 | 1 | 15 | |
| GSE54993 | YWHAB | 7529 | 0 | 17 | 53 | |
| GSE54994 | YWHAB | 7529 | 25 | 0 | 28 | |
| GSE60625 | YWHAB | 7529 | 0 | 0 | 11 | |
| GSE74703 | YWHAB | 7529 | 10 | 1 | 25 | |
| GSE74704 | YWHAB | 7529 | 9 | 0 | 11 | |
| TCGA | YWHAB | 7529 | 46 | 3 | 47 |
Total number of gains: 181; Total number of losses: 25; Total Number of normals: 282.
Somatic mutations of YWHAB:
Generating mutation plots.
Highly correlated genes for YWHAB:
Showing top 20/648 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| YWHAB | DNAJC13 | 0.760825 | 3 | 0 | 3 |
| YWHAB | PSEN1 | 0.753437 | 3 | 0 | 3 |
| YWHAB | ZNF354B | 0.726192 | 3 | 0 | 3 |
| YWHAB | FCHO2 | 0.71553 | 4 | 0 | 4 |
| YWHAB | SNX13 | 0.711403 | 3 | 0 | 3 |
| YWHAB | CDC73 | 0.70385 | 4 | 0 | 3 |
| YWHAB | GLTP | 0.701665 | 3 | 0 | 3 |
| YWHAB | KDM2B | 0.68116 | 4 | 0 | 3 |
| YWHAB | NCKAP1 | 0.679781 | 4 | 0 | 3 |
| YWHAB | ZDHHC21 | 0.678625 | 3 | 0 | 3 |
| YWHAB | STK35 | 0.677384 | 4 | 0 | 4 |
| YWHAB | ELMO2 | 0.676844 | 9 | 0 | 7 |
| YWHAB | MCEE | 0.670869 | 4 | 0 | 4 |
| YWHAB | GYS2 | 0.667254 | 4 | 0 | 3 |
| YWHAB | OXNAD1 | 0.666921 | 4 | 0 | 3 |
| YWHAB | ALOX12 | 0.666153 | 3 | 0 | 3 |
| YWHAB | ACADVL | 0.664447 | 4 | 0 | 4 |
| YWHAB | IL18 | 0.661988 | 4 | 0 | 3 |
| YWHAB | CNOT2 | 0.66069 | 4 | 0 | 3 |
| YWHAB | TBC1D22B | 0.659532 | 4 | 0 | 4 |
For details and further investigation, click here