| Full name: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | Alias Symbol: KCIP-1|14-3-3-zeta | ||
| Type: protein-coding gene | Cytoband: 8q22.3 | ||
| Entrez ID: 7534 | HGNC ID: HGNC:12855 | Ensembl Gene: ENSG00000164924 | OMIM ID: 601288 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
YWHAZ involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04390 | Hippo signaling pathway |
Expression of YWHAZ:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | YWHAZ | 7534 | 200640_at | 0.1012 | 0.7954 | |
| GSE20347 | YWHAZ | 7534 | 200639_s_at | -0.2899 | 0.0581 | |
| GSE23400 | YWHAZ | 7534 | 200640_at | -0.1006 | 0.1814 | |
| GSE26886 | YWHAZ | 7534 | 200639_s_at | -0.6871 | 0.0000 | |
| GSE29001 | YWHAZ | 7534 | 200639_s_at | -0.1502 | 0.3947 | |
| GSE38129 | YWHAZ | 7534 | 200639_s_at | -0.1003 | 0.5610 | |
| GSE45670 | YWHAZ | 7534 | 200639_s_at | 0.2000 | 0.0473 | |
| GSE53622 | YWHAZ | 7534 | 47129 | 0.0627 | 0.5961 | |
| GSE53624 | YWHAZ | 7534 | 47129 | -0.0857 | 0.1584 | |
| GSE63941 | YWHAZ | 7534 | 200638_s_at | 0.4181 | 0.1735 | |
| GSE77861 | YWHAZ | 7534 | 200639_s_at | -0.0585 | 0.5929 | |
| GSE97050 | YWHAZ | 7534 | A_32_P198923 | 0.1377 | 0.5222 | |
| SRP007169 | YWHAZ | 7534 | RNAseq | -1.2668 | 0.0003 | |
| SRP008496 | YWHAZ | 7534 | RNAseq | -1.0965 | 0.0000 | |
| SRP064894 | YWHAZ | 7534 | RNAseq | -0.2269 | 0.2950 | |
| SRP133303 | YWHAZ | 7534 | RNAseq | -0.2200 | 0.1819 | |
| SRP159526 | YWHAZ | 7534 | RNAseq | -0.3781 | 0.3490 | |
| SRP193095 | YWHAZ | 7534 | RNAseq | -0.5759 | 0.0001 | |
| SRP219564 | YWHAZ | 7534 | RNAseq | -0.6412 | 0.1178 | |
| TCGA | YWHAZ | 7534 | RNAseq | 0.1949 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
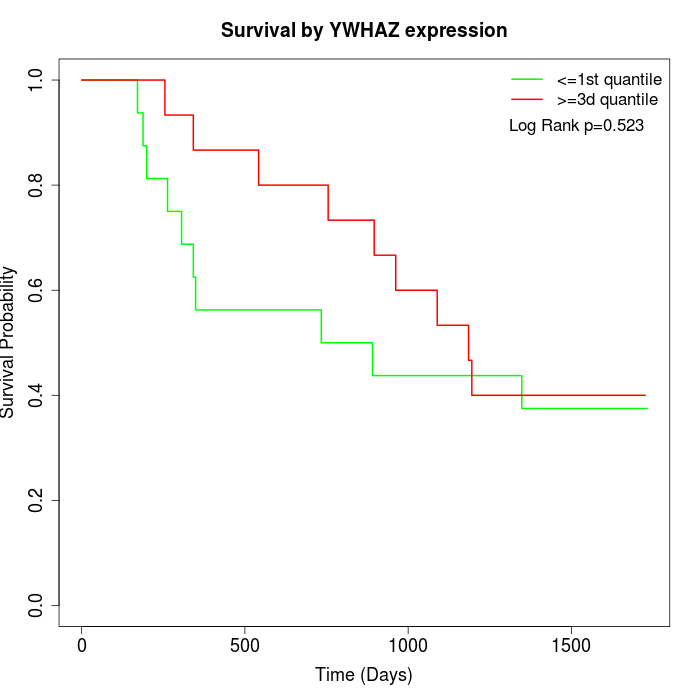
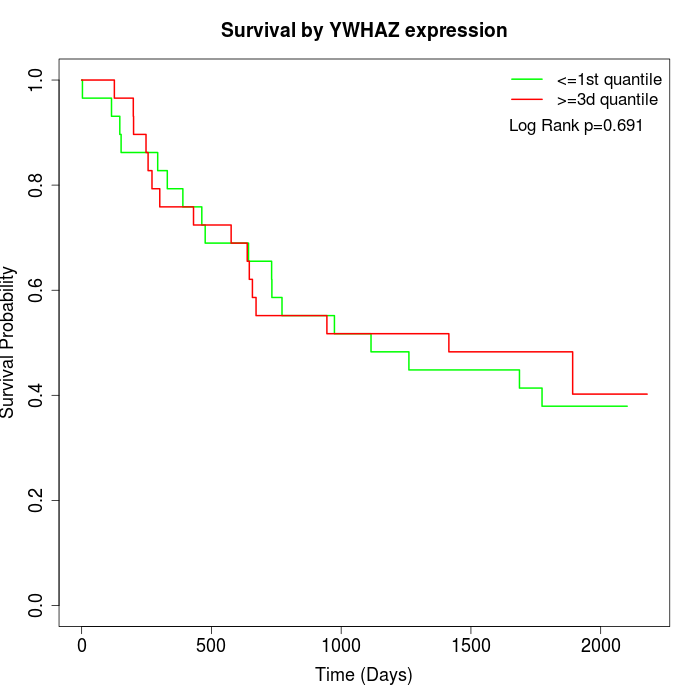
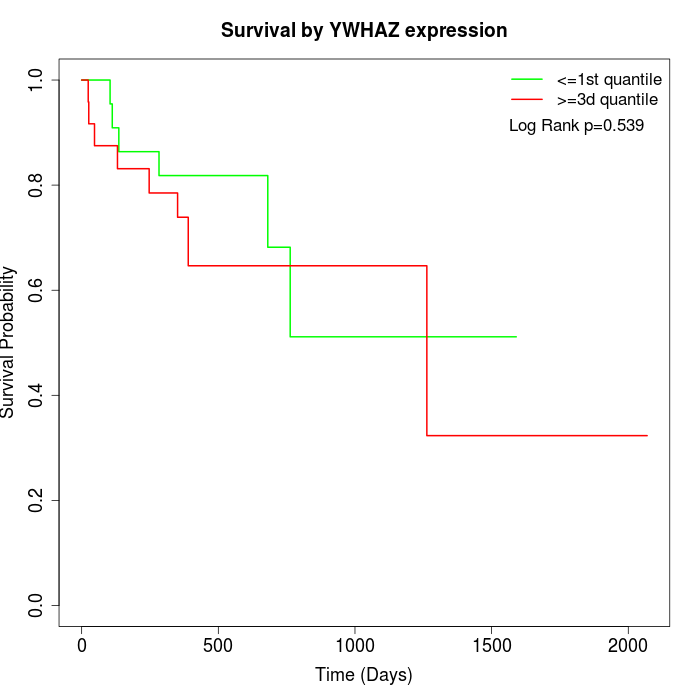
Survival by YWHAZ expression:
Note: Click image to view full size file.
Copy number change of YWHAZ:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | YWHAZ | 7534 | 17 | 0 | 13 | |
| GSE20123 | YWHAZ | 7534 | 17 | 0 | 13 | |
| GSE43470 | YWHAZ | 7534 | 22 | 0 | 21 | |
| GSE46452 | YWHAZ | 7534 | 24 | 0 | 35 | |
| GSE47630 | YWHAZ | 7534 | 24 | 0 | 16 | |
| GSE54993 | YWHAZ | 7534 | 0 | 20 | 50 | |
| GSE54994 | YWHAZ | 7534 | 38 | 1 | 14 | |
| GSE60625 | YWHAZ | 7534 | 0 | 4 | 7 | |
| GSE74703 | YWHAZ | 7534 | 19 | 0 | 17 | |
| GSE74704 | YWHAZ | 7534 | 12 | 0 | 8 | |
| TCGA | YWHAZ | 7534 | 59 | 1 | 36 |
Total number of gains: 232; Total number of losses: 26; Total Number of normals: 230.
Somatic mutations of YWHAZ:
Generating mutation plots.
Highly correlated genes for YWHAZ:
Showing top 20/393 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| YWHAZ | SRD5A3 | 0.674766 | 3 | 0 | 3 |
| YWHAZ | RIPK3 | 0.66944 | 4 | 0 | 4 |
| YWHAZ | MAL2 | 0.662354 | 4 | 0 | 4 |
| YWHAZ | SEMA5A | 0.662181 | 3 | 0 | 3 |
| YWHAZ | MTHFR | 0.656269 | 3 | 0 | 3 |
| YWHAZ | EMP1 | 0.639443 | 4 | 0 | 3 |
| YWHAZ | LRG1 | 0.639254 | 3 | 0 | 3 |
| YWHAZ | RAB22A | 0.638429 | 5 | 0 | 4 |
| YWHAZ | SLC35F5 | 0.637378 | 5 | 0 | 4 |
| YWHAZ | ZNF706 | 0.634528 | 7 | 0 | 6 |
| YWHAZ | FAM126B | 0.632239 | 4 | 0 | 3 |
| YWHAZ | ARF6 | 0.631228 | 8 | 0 | 7 |
| YWHAZ | CPNE3 | 0.623093 | 8 | 0 | 7 |
| YWHAZ | PCAT6 | 0.61832 | 4 | 0 | 3 |
| YWHAZ | TWF1 | 0.618018 | 9 | 0 | 6 |
| YWHAZ | PERP | 0.614067 | 9 | 0 | 7 |
| YWHAZ | SLC25A43 | 0.613064 | 5 | 0 | 3 |
| YWHAZ | UNC5B | 0.610343 | 6 | 0 | 4 |
| YWHAZ | OSBPL2 | 0.609132 | 5 | 0 | 3 |
| YWHAZ | SERINC2 | 0.607419 | 5 | 0 | 4 |
For details and further investigation, click here