| Full name: zinc finger protein 660 | Alias Symbol: FLJ36870 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 285349 | HGNC ID: HGNC:26720 | Ensembl Gene: ENSG00000144792 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ZNF660:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZNF660 | 285349 | 1561361_at | -0.0260 | 0.9187 | |
| GSE26886 | ZNF660 | 285349 | 1561361_at | 0.0299 | 0.7616 | |
| GSE45670 | ZNF660 | 285349 | 1561361_at | -0.1271 | 0.1554 | |
| GSE53622 | ZNF660 | 285349 | 23748 | -0.4943 | 0.0003 | |
| GSE53624 | ZNF660 | 285349 | 23748 | -0.4294 | 0.0002 | |
| GSE63941 | ZNF660 | 285349 | 1561361_at | -0.8204 | 0.0001 | |
| GSE77861 | ZNF660 | 285349 | 1561361_at | -0.0029 | 0.9814 | |
| GSE97050 | ZNF660 | 285349 | A_32_P375286 | -0.2608 | 0.3560 | |
| SRP007169 | ZNF660 | 285349 | RNAseq | 0.9660 | 0.1771 | |
| SRP064894 | ZNF660 | 285349 | RNAseq | -0.1796 | 0.4471 | |
| SRP133303 | ZNF660 | 285349 | RNAseq | -0.2714 | 0.0322 | |
| SRP159526 | ZNF660 | 285349 | RNAseq | -0.4193 | 0.1835 | |
| SRP193095 | ZNF660 | 285349 | RNAseq | 0.1962 | 0.1377 | |
| SRP219564 | ZNF660 | 285349 | RNAseq | -0.2604 | 0.6279 | |
| TCGA | ZNF660 | 285349 | RNAseq | -1.2582 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 1.
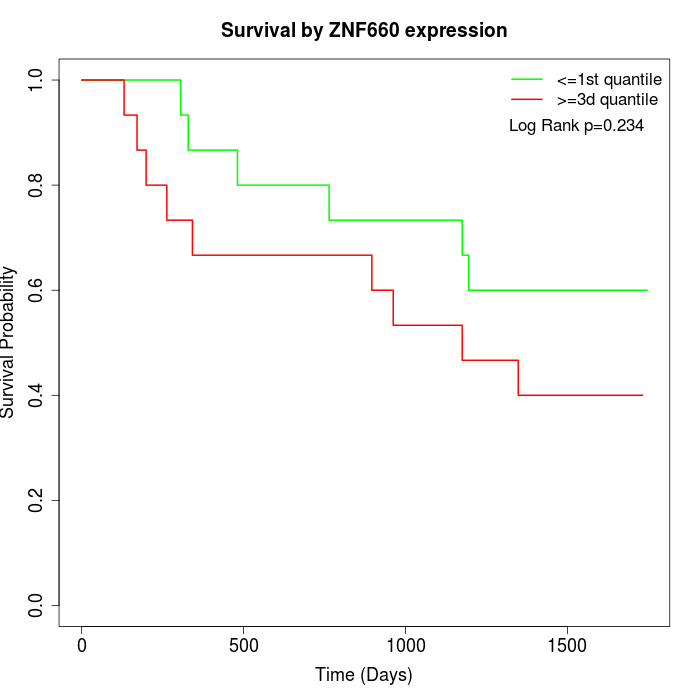
Survival by ZNF660 expression:
Note: Click image to view full size file.
Copy number change of ZNF660:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZNF660 | 285349 | 0 | 17 | 13 | |
| GSE20123 | ZNF660 | 285349 | 0 | 18 | 12 | |
| GSE43470 | ZNF660 | 285349 | 0 | 21 | 22 | |
| GSE46452 | ZNF660 | 285349 | 2 | 17 | 40 | |
| GSE47630 | ZNF660 | 285349 | 1 | 24 | 15 | |
| GSE54993 | ZNF660 | 285349 | 7 | 2 | 61 | |
| GSE54994 | ZNF660 | 285349 | 0 | 35 | 18 | |
| GSE60625 | ZNF660 | 285349 | 5 | 0 | 6 | |
| GSE74703 | ZNF660 | 285349 | 0 | 16 | 20 | |
| GSE74704 | ZNF660 | 285349 | 0 | 13 | 7 | |
| TCGA | ZNF660 | 285349 | 1 | 74 | 21 |
Total number of gains: 16; Total number of losses: 237; Total Number of normals: 235.
Somatic mutations of ZNF660:
Generating mutation plots.
Highly correlated genes for ZNF660:
Showing top 20/520 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZNF660 | PCDH9 | 0.814788 | 3 | 0 | 3 |
| ZNF660 | AHI1 | 0.744837 | 3 | 0 | 3 |
| ZNF660 | ABCA6 | 0.721031 | 5 | 0 | 5 |
| ZNF660 | ADAMTS5 | 0.718875 | 5 | 0 | 5 |
| ZNF660 | VAT1L | 0.718177 | 5 | 0 | 5 |
| ZNF660 | FIBIN | 0.712088 | 3 | 0 | 3 |
| ZNF660 | GALNT15 | 0.711325 | 3 | 0 | 3 |
| ZNF660 | SMAD9 | 0.706199 | 5 | 0 | 5 |
| ZNF660 | ITPKB | 0.706003 | 3 | 0 | 3 |
| ZNF660 | KATNAL1 | 0.704313 | 4 | 0 | 4 |
| ZNF660 | SGCG | 0.695503 | 5 | 0 | 5 |
| ZNF660 | TMEM71 | 0.691928 | 4 | 0 | 4 |
| ZNF660 | WFS1 | 0.691071 | 4 | 0 | 3 |
| ZNF660 | JAZF1 | 0.690436 | 4 | 0 | 4 |
| ZNF660 | MEOX2 | 0.688686 | 4 | 0 | 4 |
| ZNF660 | TOX | 0.688554 | 4 | 0 | 4 |
| ZNF660 | ZFPM2 | 0.685081 | 5 | 0 | 5 |
| ZNF660 | RHOQ | 0.684237 | 3 | 0 | 3 |
| ZNF660 | GABARAPL1 | 0.682468 | 4 | 0 | 3 |
| ZNF660 | TRAK2 | 0.680736 | 5 | 0 | 4 |
For details and further investigation, click here