| Full name: acyl-CoA dehydrogenase medium chain | Alias Symbol: MCAD|MCADH|ACAD1 | ||
| Type: protein-coding gene | Cytoband: 1p31.1 | ||
| Entrez ID: 34 | HGNC ID: HGNC:89 | Ensembl Gene: ENSG00000117054 | OMIM ID: 607008 |
| Drug and gene relationship at DGIdb | |||
ACADM involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway |
Expression of ACADM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACADM | 34 | 202502_at | -0.8880 | 0.0674 | |
| GSE20347 | ACADM | 34 | 202502_at | -1.4752 | 0.0000 | |
| GSE23400 | ACADM | 34 | 202502_at | -1.2216 | 0.0000 | |
| GSE26886 | ACADM | 34 | 202502_at | -1.9422 | 0.0000 | |
| GSE29001 | ACADM | 34 | 202502_at | -1.4507 | 0.0033 | |
| GSE38129 | ACADM | 34 | 202502_at | -1.2578 | 0.0000 | |
| GSE45670 | ACADM | 34 | 202502_at | -0.7581 | 0.0142 | |
| GSE53622 | ACADM | 34 | 88593 | -1.2863 | 0.0000 | |
| GSE53624 | ACADM | 34 | 88593 | -1.4891 | 0.0000 | |
| GSE63941 | ACADM | 34 | 202502_at | 0.3677 | 0.4085 | |
| GSE77861 | ACADM | 34 | 202502_at | -1.9457 | 0.0002 | |
| GSE97050 | ACADM | 34 | A_23_P96761 | -1.3341 | 0.1138 | |
| SRP007169 | ACADM | 34 | RNAseq | -3.1744 | 0.0000 | |
| SRP008496 | ACADM | 34 | RNAseq | -2.9921 | 0.0000 | |
| SRP064894 | ACADM | 34 | RNAseq | -1.9588 | 0.0000 | |
| SRP133303 | ACADM | 34 | RNAseq | -1.2239 | 0.0000 | |
| SRP159526 | ACADM | 34 | RNAseq | -1.9273 | 0.0000 | |
| SRP193095 | ACADM | 34 | RNAseq | -1.8348 | 0.0000 | |
| SRP219564 | ACADM | 34 | RNAseq | -1.6619 | 0.0002 | |
| TCGA | ACADM | 34 | RNAseq | -0.3608 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 15.
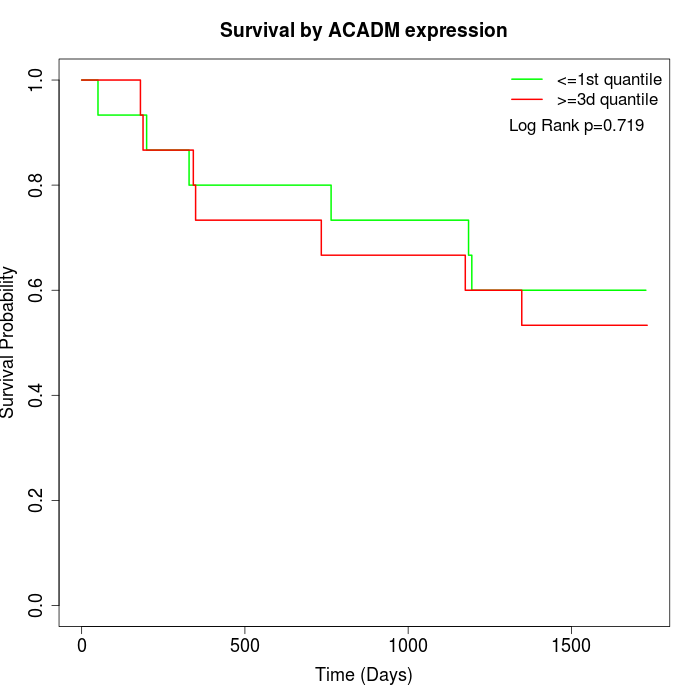
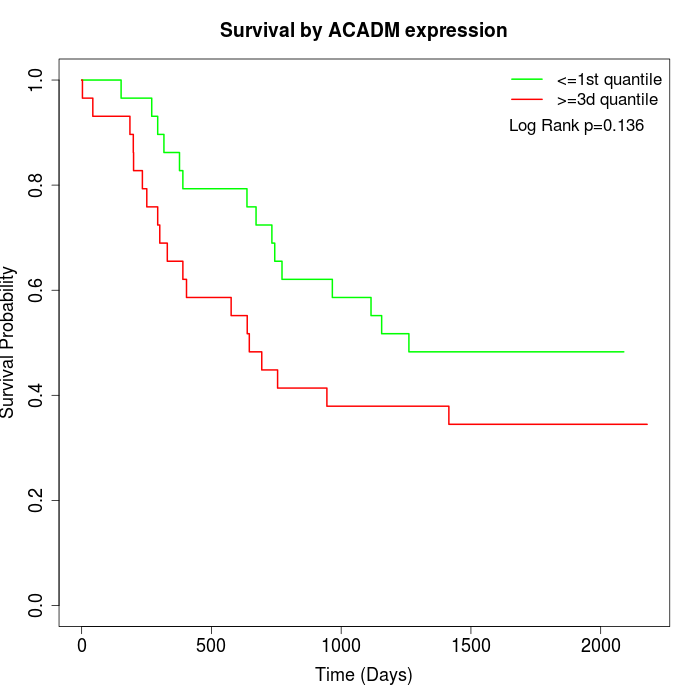
Survival by ACADM expression:
Note: Click image to view full size file.
Copy number change of ACADM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACADM | 34 | 0 | 7 | 23 | |
| GSE20123 | ACADM | 34 | 1 | 7 | 22 | |
| GSE43470 | ACADM | 34 | 4 | 4 | 35 | |
| GSE46452 | ACADM | 34 | 0 | 1 | 58 | |
| GSE47630 | ACADM | 34 | 8 | 6 | 26 | |
| GSE54993 | ACADM | 34 | 0 | 0 | 70 | |
| GSE54994 | ACADM | 34 | 6 | 3 | 44 | |
| GSE60625 | ACADM | 34 | 0 | 0 | 11 | |
| GSE74703 | ACADM | 34 | 3 | 3 | 30 | |
| GSE74704 | ACADM | 34 | 1 | 4 | 15 | |
| TCGA | ACADM | 34 | 8 | 24 | 64 |
Total number of gains: 31; Total number of losses: 59; Total Number of normals: 398.
Somatic mutations of ACADM:
Generating mutation plots.
Highly correlated genes for ACADM:
Showing top 20/1753 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACADM | VSIG2 | 0.813593 | 5 | 0 | 5 |
| ACADM | MPP7 | 0.809813 | 7 | 0 | 7 |
| ACADM | KAT2B | 0.808486 | 11 | 0 | 11 |
| ACADM | FCHO2 | 0.807007 | 8 | 0 | 7 |
| ACADM | LMBRD1 | 0.79953 | 11 | 0 | 11 |
| ACADM | CCNG2 | 0.789725 | 11 | 0 | 11 |
| ACADM | CA13 | 0.788206 | 3 | 0 | 3 |
| ACADM | FAM214A | 0.779553 | 6 | 0 | 6 |
| ACADM | ZNRF1 | 0.77833 | 3 | 0 | 3 |
| ACADM | EMC3 | 0.776745 | 10 | 0 | 10 |
| ACADM | CAST | 0.775338 | 11 | 0 | 11 |
| ACADM | TAB3 | 0.775005 | 6 | 0 | 6 |
| ACADM | SORT1 | 0.77346 | 11 | 0 | 10 |
| ACADM | RANBP9 | 0.768719 | 11 | 0 | 10 |
| ACADM | CPEB3 | 0.768476 | 10 | 0 | 10 |
| ACADM | SFTA2 | 0.768283 | 4 | 0 | 4 |
| ACADM | HPGD | 0.768143 | 11 | 0 | 10 |
| ACADM | ELMO2 | 0.768004 | 10 | 0 | 10 |
| ACADM | PPP1R3C | 0.76762 | 11 | 0 | 11 |
| ACADM | ETFDH | 0.76739 | 10 | 0 | 9 |
For details and further investigation, click here