| Full name: acyl-CoA dehydrogenase short chain | Alias Symbol: SCAD|ACAD3 | ||
| Type: protein-coding gene | Cytoband: 12q24.31 | ||
| Entrez ID: 35 | HGNC ID: HGNC:90 | Ensembl Gene: ENSG00000122971 | OMIM ID: 606885 |
| Drug and gene relationship at DGIdb | |||
Expression of ACADS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACADS | 35 | 202366_at | -0.1838 | 0.4703 | |
| GSE20347 | ACADS | 35 | 202366_at | -0.0099 | 0.9348 | |
| GSE23400 | ACADS | 35 | 202366_at | -0.1287 | 0.0016 | |
| GSE26886 | ACADS | 35 | 202366_at | 0.1571 | 0.1415 | |
| GSE29001 | ACADS | 35 | 202366_at | -0.0482 | 0.8174 | |
| GSE38129 | ACADS | 35 | 202366_at | -0.2059 | 0.0434 | |
| GSE45670 | ACADS | 35 | 202366_at | -0.1038 | 0.2583 | |
| GSE53622 | ACADS | 35 | 2459 | -0.4201 | 0.0008 | |
| GSE53624 | ACADS | 35 | 2459 | -0.2290 | 0.0509 | |
| GSE63941 | ACADS | 35 | 202366_at | 0.1886 | 0.3348 | |
| GSE77861 | ACADS | 35 | 202366_at | -0.0257 | 0.8829 | |
| GSE97050 | ACADS | 35 | A_23_P65022 | -0.2793 | 0.3453 | |
| SRP007169 | ACADS | 35 | RNAseq | -0.1712 | 0.7311 | |
| SRP008496 | ACADS | 35 | RNAseq | -0.2841 | 0.4433 | |
| SRP064894 | ACADS | 35 | RNAseq | 0.3435 | 0.2853 | |
| SRP133303 | ACADS | 35 | RNAseq | -0.4096 | 0.0439 | |
| SRP159526 | ACADS | 35 | RNAseq | -0.1226 | 0.6553 | |
| SRP193095 | ACADS | 35 | RNAseq | -0.7554 | 0.0015 | |
| SRP219564 | ACADS | 35 | RNAseq | -0.3301 | 0.5177 | |
| TCGA | ACADS | 35 | RNAseq | -0.5789 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
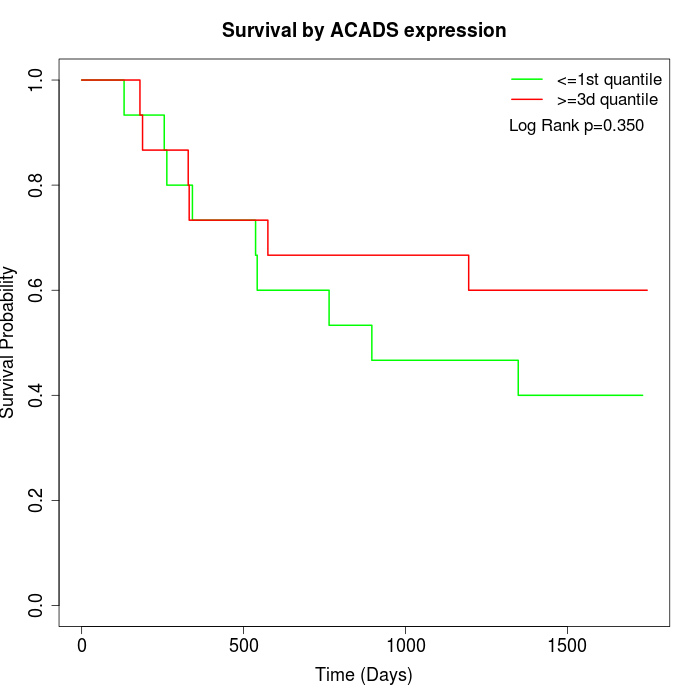
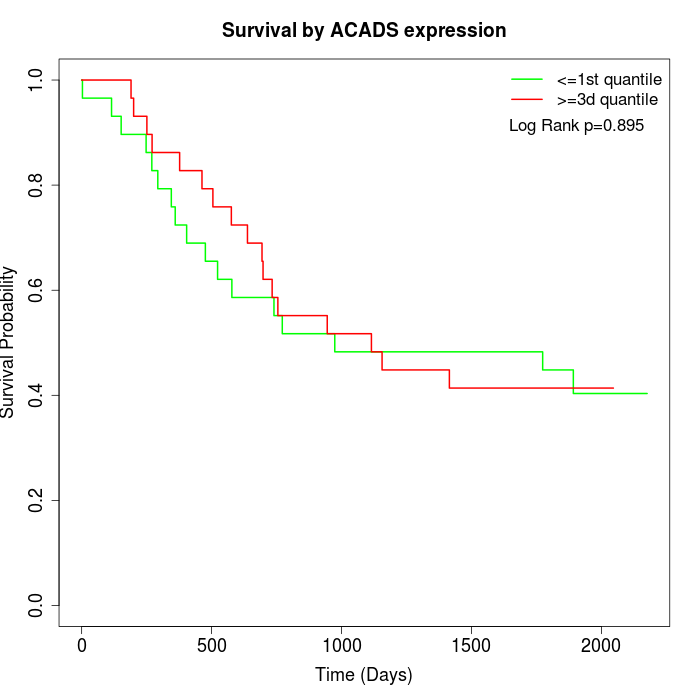
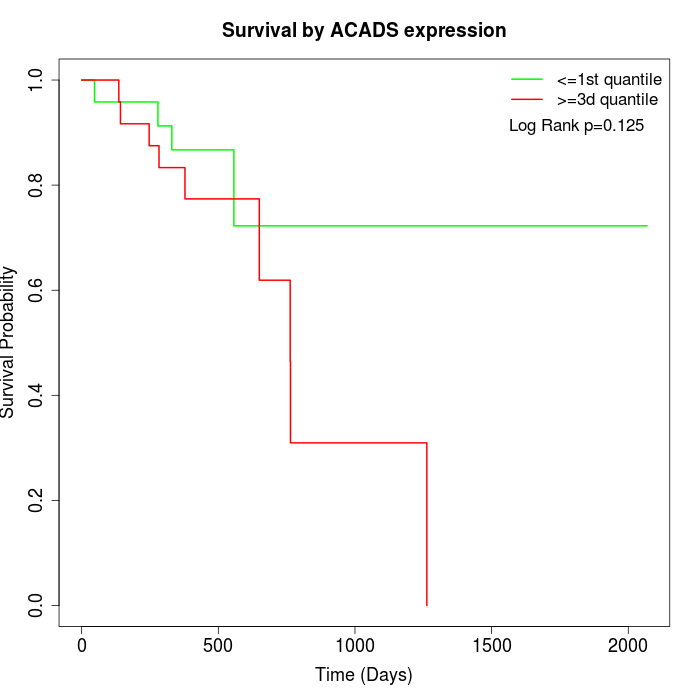
Survival by ACADS expression:
Note: Click image to view full size file.
Copy number change of ACADS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACADS | 35 | 6 | 2 | 22 | |
| GSE20123 | ACADS | 35 | 6 | 2 | 22 | |
| GSE43470 | ACADS | 35 | 2 | 2 | 39 | |
| GSE46452 | ACADS | 35 | 9 | 1 | 49 | |
| GSE47630 | ACADS | 35 | 9 | 3 | 28 | |
| GSE54993 | ACADS | 35 | 0 | 5 | 65 | |
| GSE54994 | ACADS | 35 | 7 | 6 | 40 | |
| GSE60625 | ACADS | 35 | 0 | 0 | 11 | |
| GSE74703 | ACADS | 35 | 2 | 1 | 33 | |
| GSE74704 | ACADS | 35 | 3 | 2 | 15 | |
| TCGA | ACADS | 35 | 20 | 11 | 65 |
Total number of gains: 64; Total number of losses: 35; Total Number of normals: 389.
Somatic mutations of ACADS:
Generating mutation plots.
Highly correlated genes for ACADS:
Showing top 20/310 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACADS | TEF | 0.768317 | 3 | 0 | 3 |
| ACADS | ASTN1 | 0.759753 | 3 | 0 | 3 |
| ACADS | ADCY1 | 0.70374 | 3 | 0 | 3 |
| ACADS | CDC42EP2 | 0.688236 | 4 | 0 | 3 |
| ACADS | ADAMTSL3 | 0.685841 | 4 | 0 | 3 |
| ACADS | KBTBD12 | 0.669975 | 4 | 0 | 3 |
| ACADS | ADRA2B | 0.66923 | 3 | 0 | 3 |
| ACADS | AGAP2 | 0.666778 | 4 | 0 | 4 |
| ACADS | PDCD1 | 0.665766 | 3 | 0 | 3 |
| ACADS | C4BPA | 0.656821 | 3 | 0 | 3 |
| ACADS | ACTR10 | 0.654007 | 3 | 0 | 3 |
| ACADS | ZFHX2 | 0.651433 | 4 | 0 | 4 |
| ACADS | SLC6A13 | 0.649383 | 5 | 0 | 5 |
| ACADS | SCGN | 0.64914 | 4 | 0 | 4 |
| ACADS | ABCB1 | 0.647865 | 3 | 0 | 3 |
| ACADS | HRC | 0.646773 | 5 | 0 | 5 |
| ACADS | HRH3 | 0.642587 | 4 | 0 | 3 |
| ACADS | MOB2 | 0.641119 | 5 | 0 | 3 |
| ACADS | FERD3L | 0.64048 | 4 | 0 | 3 |
| ACADS | NKAIN1 | 0.639796 | 3 | 0 | 3 |
For details and further investigation, click here