| Full name: DNA polymerase epsilon 3, accessory subunit | Alias Symbol: CHRAC17|Ybl1|p17|CHARAC17|CHRAC2 | ||
| Type: protein-coding gene | Cytoband: 9q32 | ||
| Entrez ID: 54107 | HGNC ID: HGNC:13546 | Ensembl Gene: ENSG00000148229 | OMIM ID: 607267 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
POLE3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05166 | HTLV-I infection |
Expression of POLE3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | POLE3 | 54107 | 208828_at | 0.0775 | 0.9011 | |
| GSE20347 | POLE3 | 54107 | 208828_at | -0.0358 | 0.9086 | |
| GSE23400 | POLE3 | 54107 | 208828_at | 0.8213 | 0.0000 | |
| GSE26886 | POLE3 | 54107 | 208828_at | -1.0404 | 0.0003 | |
| GSE29001 | POLE3 | 54107 | 208828_at | 0.4597 | 0.1672 | |
| GSE38129 | POLE3 | 54107 | 208828_at | 0.3983 | 0.0364 | |
| GSE45670 | POLE3 | 54107 | 208828_at | 0.3736 | 0.0079 | |
| GSE53622 | POLE3 | 54107 | 8933 | 0.3852 | 0.0000 | |
| GSE53624 | POLE3 | 54107 | 8933 | 0.6072 | 0.0000 | |
| GSE63941 | POLE3 | 54107 | 208828_at | 1.6043 | 0.0000 | |
| GSE77861 | POLE3 | 54107 | 208828_at | 0.2386 | 0.4250 | |
| GSE97050 | POLE3 | 54107 | A_23_P135326 | 0.1672 | 0.4985 | |
| SRP007169 | POLE3 | 54107 | RNAseq | 1.6217 | 0.0002 | |
| SRP008496 | POLE3 | 54107 | RNAseq | 1.4934 | 0.0000 | |
| SRP064894 | POLE3 | 54107 | RNAseq | 0.5342 | 0.0015 | |
| SRP133303 | POLE3 | 54107 | RNAseq | 0.5837 | 0.0026 | |
| SRP159526 | POLE3 | 54107 | RNAseq | 0.5225 | 0.0176 | |
| SRP193095 | POLE3 | 54107 | RNAseq | 0.2202 | 0.1949 | |
| SRP219564 | POLE3 | 54107 | RNAseq | 0.2570 | 0.4497 | |
| TCGA | POLE3 | 54107 | RNAseq | 0.2662 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 1.
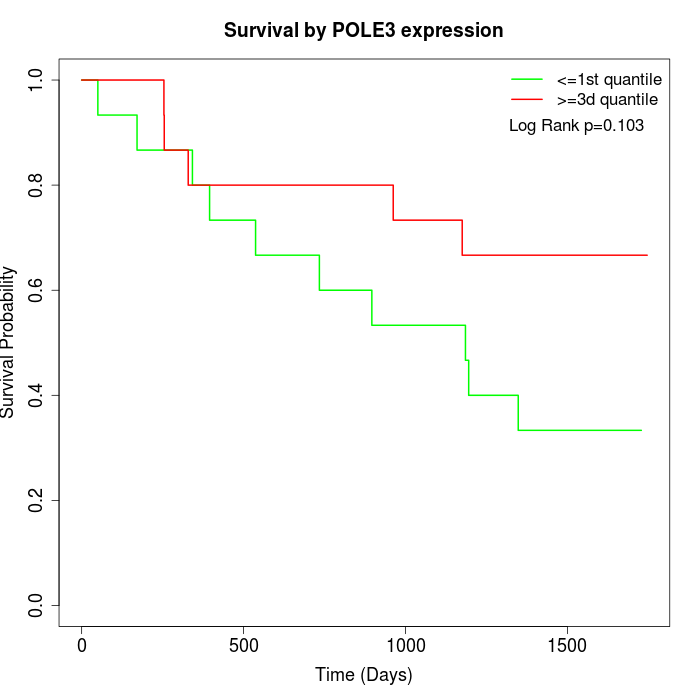
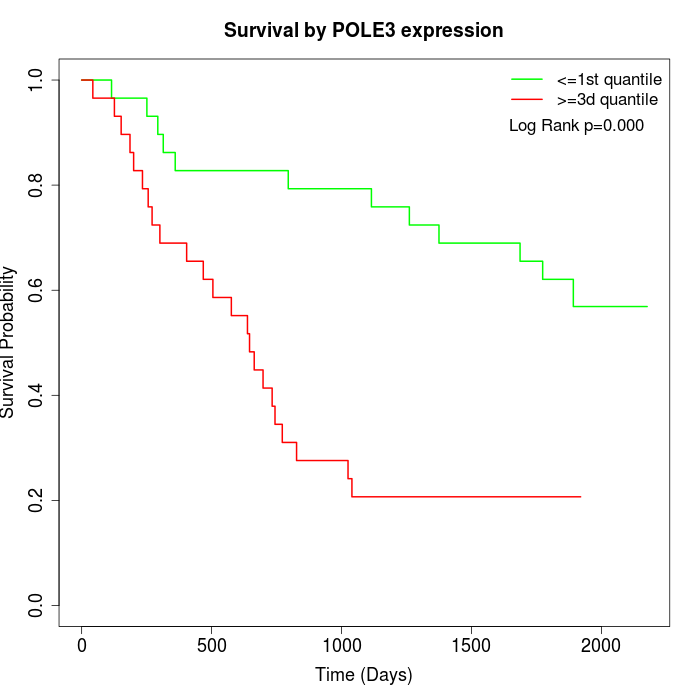
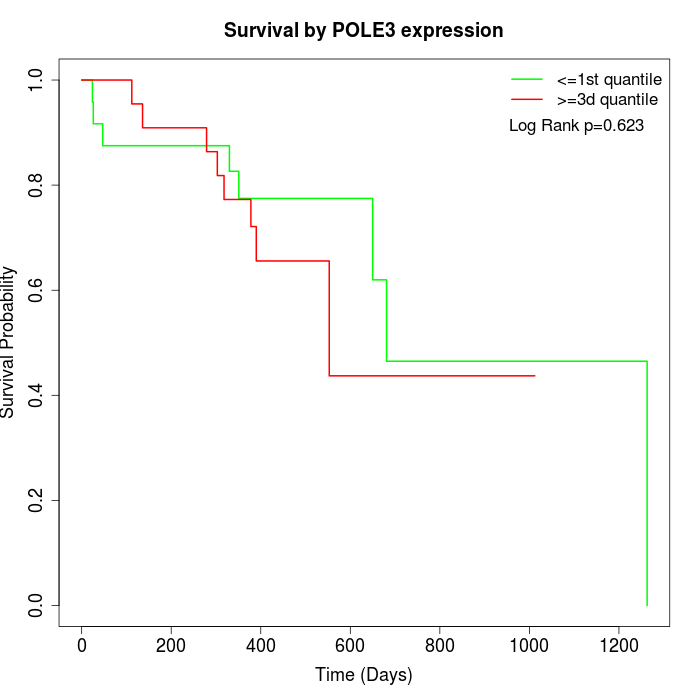
Survival by POLE3 expression:
Note: Click image to view full size file.
Copy number change of POLE3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | POLE3 | 54107 | 4 | 10 | 16 | |
| GSE20123 | POLE3 | 54107 | 4 | 10 | 16 | |
| GSE43470 | POLE3 | 54107 | 4 | 3 | 36 | |
| GSE46452 | POLE3 | 54107 | 6 | 13 | 40 | |
| GSE47630 | POLE3 | 54107 | 3 | 16 | 21 | |
| GSE54993 | POLE3 | 54107 | 3 | 3 | 64 | |
| GSE54994 | POLE3 | 54107 | 9 | 10 | 34 | |
| GSE60625 | POLE3 | 54107 | 0 | 0 | 11 | |
| GSE74703 | POLE3 | 54107 | 4 | 3 | 29 | |
| GSE74704 | POLE3 | 54107 | 2 | 8 | 10 | |
| TCGA | POLE3 | 54107 | 26 | 22 | 48 |
Total number of gains: 65; Total number of losses: 98; Total Number of normals: 325.
Somatic mutations of POLE3:
Generating mutation plots.
Highly correlated genes for POLE3:
Showing top 20/957 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| POLE3 | SPRYD4 | 0.827588 | 3 | 0 | 3 |
| POLE3 | PRPF38A | 0.806682 | 4 | 0 | 4 |
| POLE3 | AP1M2 | 0.801593 | 4 | 0 | 4 |
| POLE3 | EAF1 | 0.789014 | 3 | 0 | 3 |
| POLE3 | MCU | 0.777932 | 3 | 0 | 3 |
| POLE3 | TPD52 | 0.774639 | 3 | 0 | 3 |
| POLE3 | PANK1 | 0.758785 | 3 | 0 | 3 |
| POLE3 | TP53RK | 0.756729 | 3 | 0 | 3 |
| POLE3 | CD163L1 | 0.756534 | 3 | 0 | 3 |
| POLE3 | DHX38 | 0.755074 | 3 | 0 | 3 |
| POLE3 | SREBF2 | 0.737495 | 3 | 0 | 3 |
| POLE3 | ACAP2 | 0.732605 | 3 | 0 | 3 |
| POLE3 | TRMT1L | 0.728614 | 3 | 0 | 3 |
| POLE3 | CERS3 | 0.719001 | 3 | 0 | 3 |
| POLE3 | LCMT1 | 0.713779 | 3 | 0 | 3 |
| POLE3 | GABPA | 0.713423 | 4 | 0 | 4 |
| POLE3 | AKT1S1 | 0.70963 | 3 | 0 | 3 |
| POLE3 | TMEM117 | 0.707309 | 4 | 0 | 4 |
| POLE3 | KLF3 | 0.706555 | 3 | 0 | 3 |
| POLE3 | SLC15A2 | 0.704767 | 3 | 0 | 3 |
For details and further investigation, click here